Abstract
Purpose:
This study examined the utility of sets of single-nucleotide polymorphisms (SNPs) in familial but non-BRCA-associated breast cancer (BC).
Methods:
We derived a polygenic risk score (PRS) based on 24 known BC risk SNPs for 4,365 women from the Breast Cancer Family Registry and Kathleen Cuningham Consortium Foundation for Research into Familial Breast Cancer familial BC cohorts. We compared scores for women based on cancer status at baseline; 2,599 women unaffected at enrollment were followed-up for an average of 7.4 years. Cox proportional hazards regression was used to analyze the association of PRS with BC risk. The BOADICEA risk prediction algorithm was used to measure risk based on family history alone.
Results:
The mean PRS at baseline was 2.25 (SD, 0.35) for affected women and was 2.17 (SD, 0.35) for unaffected women from combined cohorts (P < 10–6). During follow-up, 205 BC cases occurred. The hazard ratios for continuous PRS (per SD) and upper versus lower quintiles were 1.38 (95% confidence interval: 1.22–1.56) and 3.18 (95% confidence interval: 1.84–5.23) respectively. Based on their PRS-based predicted risk, management for up to 23% of women could be altered.
Conclusion:
Including BC-associated SNPs in risk assessment can provide more accurate risk prediction than family history alone and can influence recommendations for cancer screening and prevention modalities for high-risk women.
Genet Med 19 1, 30–35.
Similar content being viewed by others
Introduction
In the recent initiative toward “precision medicine” announced by the National Institutes of Health,1 the use of genetic information for the identification of high-risk groups for targeted screening and/or prevention is gradually becoming part of routine medical care. Once limited to pathogenic mutations in high-risk genes such as BRCA1, BRCA2, p53, and the mismatch repair genes associated with Lynch syndrome, the past decade has seen the identification of additional genes for which pathogenic variants are associated with perhaps two- to five-fold increased risks of cancer, as well as an ever-increasing set of common single-nucleotide polymorphisms (SNPs), each of which is associated with a relative risk of 1.05 to 1.3 of developing breast cancer.2,3 Although these SNPs are not useful for risk prediction when considered individually, theoretical calculations indicate that a combined score based on genotypes in a large number of such loci could have substantial predictive value for risk stratification in the general population4,5 as well as in BRCA1 and BRCA2 carriers.6 The combination of high-risk genes such as BRCA1 and BRCA2 and the known SNPs described here is estimated to explain less than half of the familial aggregation of breast cancer. This notwithstanding, such sets of SNPs may have clinically useful predictive power in the familial setting due to the increased risk of breast cancer conferred by a woman’s family history alone. To date, only a single study7 has examined the utility of such SNP panels in the familial context, and none have examined it in a prospective fashion. Sawyer et al.7 looked at differences in a PRS based on 22 SNPs between BRCA1/2 carriers and BRCA1/2-negative women with breast cancer from a familial cancer clinic in Australia and a set of controls. They found that noncarrier cases had a higher PRS than BRCA1/2 carriers, and that a higher proportion of individuals with non-BRCA1/2 cases with a PRS in the top quartile had breast cancer diagnosed before age 30 compared with the lowest quartile. The goal of the present study was to examine the utility of panels of SNPs in the context of familial breast cancer, in which women are already at elevated risk owing to their family history, and to determine whether such SNP panels could stratify women into clinically useful risk groups. Currently, various advisory bodies have proposed guidelines for the use of magnetic resonance imaging (MRI) in addition to mammography for women at high risk. For example, the American Cancer Society8 proposes lifetime risk thresholds of 20–25% for MRI, whereas the UK NICE guidelines9 use a threshold of 30%. Here, we examined women in families not known to have BRCA1/2 mutations from two familial breast cancer resources: the Breast Cancer Family Registry (BCFR) cohort and the Kathleen Cuningham Consortium Foundation for Research into Familial Breast Cancer (kConFab). This study is novel in two ways. First, it examined women who are already at increased familial risk; second, it prospectively analyzed women who were unaffected at cohort enrollment.
Materials and Methods
SNP selection and genotyping
BCFR. For BCFR subjects, a total of 24 SNPs were successfully genotyped (Supplementary Table S1 online). These correspond to the loci known to be associated with breast cancer at the start of the study and do not include the more recent loci discovered as part of the iCOGS analyses.2,3 These SNPs were genotyped using a capture-based next-generation sequencing method developed (by A.M.) specifically for this study.
kConFab. In kConFab, SNPs were genotyped in two phases using two different technologies. In the first phase, 18 SNPs were typed using iPLEX; in the second phase, an additional 90 SNPs were typed using Fluidigm technology. To have comparable scores for the two data sets in order to enable a combined analysis, we chose 24 SNPs, which were either the same SNP or in complete or strong (R2 > 0.9) linkage disequilibrium with the SNP genotyped in BCFR. Supplementary Table S1 online shows the minor allele frequency and odds ratio for the SNPs genotyped in each cohort.
Subjects
The BCFR is a National Cancer Institute–sponsored resource of familial breast cancer (http://www.bcfamilyregistry.org).10,11 It consists of more than 15,000 families enrolled since 1995 from six sites—four in the United States (Utah, northern California, New York, Philadelphia) and one each in Australia and Canada—with data collection regarding lifestyle factors, tumor histopathology, and, increasingly, genetic information. Three of the sites incorporated a clinic-based ascertainment strategy; the other three used a population-based strategy. Recruitment and genetic studies were approved by the University of Utah institutional review board and the local institutional review boards of the BCFR centers from which we received blood samples and data. Written informed consent was obtained from each participant. Families were selected for this study on the basis of availability of DNA samples in the family and age (at least one woman diagnosed with BC younger than age 60 years prior to enrollment and one or more unaffected women older than age 30 years at baseline with a DNA sample available). In total, 2,467 women were successfully genotyped for at least 20 of the 24 SNPs; of these, 96% had at least 22 valid genotypes called. After exclusion of 376 women without the required dates of birth, enrollment, and follow-up end points, 2,091 women from 707 families were included in the analyses. Of these, 991 women in 481 families who were unaffected by BC and were younger than 70 years of age at baseline were included in the prospective analyses.
The second data set analyzed in this project was based on the kConFab12 resource that has enrolled BC families since 1997 and systematically followed women every 3 years.13 Details on the resource and ascertainment criteria have been described elsewhere (http://www.kconfab.org). Subjects were selected for genotyping based solely on their phenotype at baseline, without regard for any subsequent cancers. For this study, eligibility was restricted to families with at least one family member genotyped for the SNPs of interest. Families were systematically screened for and excluded if found to contain a mutation in BRCA1, BRCA2, PALB2, or ATM. In this study we included 2,732 women from 535 families who had sufficient genotype data to compute PRS. After exclusion of women who did not meet the inclusion criteria, 2,274 women from 523 families were eligible for analysis. Of these, 1,608 women from 488 families were included in the prospective cohort based on the same inclusion criteria as described for BCFR. All participants in this study provided signed informed consent, and the study was approved by the Human Research Ethics Committee of the Peter MacCallum Cancer Center as well as by all participating centers.
Statistical Methods
Calculation of PRS. We created a PRS for each genotyped individual based on her genotype at each of the 24 loci, defined for the  individual as
individual as  , where
, where  is the number of risk alleles carried by the
is the number of risk alleles carried by the  individual at the
individual at the  SNP,
SNP,  , and
, and  is the per-allele relative risk (estimated by the per-allele odds ratio in Europeans from large published studies3) associated with the
is the per-allele relative risk (estimated by the per-allele odds ratio in Europeans from large published studies3) associated with the  SNP. When SNP genotypes were missing for an individual (maximum of four missing genotypes per individual), they were included in the overall PRS by weighting each genotype by its expectation given the MAF at that locus and their relatives’ genotypes (if any) as estimated from 10,000 replicates of the data set using the simulation program SLINK.14
SNP. When SNP genotypes were missing for an individual (maximum of four missing genotypes per individual), they were included in the overall PRS by weighting each genotype by its expectation given the MAF at that locus and their relatives’ genotypes (if any) as estimated from 10,000 replicates of the data set using the simulation program SLINK.14
For the 24 SNPs used here, the theoretical expected value of the PRS is 2.123 with a variance of 0.117, based on the odds ratios and MAF for each SNP.
Assessment of family history. As part of the Prof-SC cohort,11 the BOADICEA model15 was used to predict BC risk in more than 18,000 unaffected women from the BCFR and KConFab cohorts. Although originally designed to predict probabilities of an individual carrying a BRCA1 or BRCA2 mutation, BOADICEA also predicts a woman’s risk of breast and ovarian cancers both for the next 10 years and until age 80 (remaining lifetime risk) and has been shown to be an accurate predictor of breast cancer risk in a prospective study.16 Specifically, we used the predicted 10-year risk of BC as calculated by BOADICEA as a summary measure of each woman’s familial risk given her age and the ages/age at diagnosis and cancer (breast, ovarian, prostate, and pancreatic) status of all relatives and incorporating any available BRCA1/2 genetic testing results. Of the 2,599 women in the prospective analysis, BOADICEA scores were available for 2,457 (95%) women. Finally, we used the BOADICEA remaining lifetime risk as a baseline for modification by PRS as described below to examine lifetime-risk changes as a function of the SNP-based PRS.
Statistical analysis of PRS scores. We compared PRS scores in women who were affected versus those unaffected by BC at entry into the BCFR or kConFab cohorts. In this analysis, all women with a PRS were included without regard to history of other cancers (e.g., ovarian cancer). To adjust for the slight differences in the specific SNPs used in the two cohorts and to express the estimated hazard ratios (HRs) per SD, we normalized the PRS scores by subtracting the theoretical mean from each score and dividing by the theoretical standard deviation prior to analysis. The primary analyses were prospective; women who were unaffected by BC and who had not undergone bilateral prophylactic mastectomy prior to cohort enrollment were eligible for follow-up with the primary end-point development of invasive BC or DCIS during the follow-up period. Women were censored by whichever of the following occurred first: (i) diagnosis of BC (invasive or DCIS), (ii) bilateral prophylactic mastectomy, (iii) death, or (iv) last follow-up questionnaire (or last date known to be alive and cancer-free). The characteristics of the 2,599 women who form the prospective cohort are presented in Table 1 . We used Cox proportional hazards models to evaluate the effect of PRS on BC risk in this cohort. In these analyses, we used the continuous PRS score as an independent predictor as well as a comparison of the upper and lower quintile of such scores (calculated separately for BCFR and kConFab cohorts). The main analyses were stratified by study center (the six BCFR sites and kConFab), and all analyses used a robust variance estimator based on family membership to adjust the variance for correlations in scores and overall cancer risks in related individuals. Interactions with family history, age, and study center were performed using multivariable Cox models including main effects and an interaction term.
To examine the effect of the PRS on the estimated lifetime risk of breast cancer as assessed by BOADICEA ( ), we estimated a SNP-based cumulative risk for each woman in the sample by
), we estimated a SNP-based cumulative risk for each woman in the sample by  , where
, where  ,
,  is the natural logarithm of the estimated HR for continuous PRS in the prospective cohort, and
is the natural logarithm of the estimated HR for continuous PRS in the prospective cohort, and  is the standardized PRS for the
is the standardized PRS for the  woman in the cohort.
woman in the cohort.
All statistical analyses were performed using STATA 12.0 (StataCorp, College Station, TX).
Results
We first compared the PRS in all subjects at baseline. A total of 1,496 women affected by breast cancer (1,084 BCFR; 412 kConFab) and 2,869 (1,007 BCFR; 1,862 kConFab) unaffected women were available for analysis. There were highly significant differences between the mean PRS in affected women at baseline compared with unaffected women in each cohort as well as in the combined set (P = 3 × 10−5, 1 × 10−6, and 1 × 10−10, respectively). The mean PRS for unaffected women of 2.170 is slightly higher than the theoretical mean of 2.123, which is expected given their selection for a positive family history. PRS scores were quite comparable between the two cohorts, especially in unaffected women. Table 1 shows the characteristics of the prospective cohort. The overall breast cancer incidence was higher in the BCFR cohort (P = 0.0012), probably because women in the BCFR were, on average, older at the start of follow-up than those in kConFab (46.4 vs. 42.6; P < 10−5) and may have had a less stringent family-history criterion for entry than that for the BCFR. The results of the Cox proportional hazards models in the analysis of prospective data are shown in Table 2 . In both of the cohorts and for both the continuous and upper versus lower quintile PRS scores, the PRS was associated with a highly significant increased risk, with an HR for upper versus lower quintile of 3.18. HRs by quintile for each study are shown in Supplementary Table S2 online. The HRs for the continuous PRS were not significantly different between the BCFR and kConFab study cohorts (P = 0.13) for study × PRS interaction but were borderline significant for the upper versus lower quintile (P = 0.05), and the HR did not vary significantly as a function of age at baseline (P = 0.88 and P = 0.71 for the two cohorts, respectively). We tested the validity of the proportional hazards assumption implicit in the Cox models; neither the quintiles defined by PRS (P = 0.85) nor the continuous PRS score (P = 0.64) showed departure from the proportional hazards assumption. In a sensitivity analyses, we excluded women who had been affected by any cancer at baseline (including ovarian) and censored women at date of diagnosis of any nonbreast cancer occurring during follow-up. Results were only slightly changed.
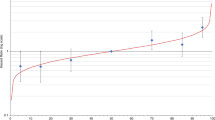
We used Kaplan–Meier survival analysis to examine the cumulative risks of BC for the lower quintile, three middle quintiles, and upper quintile, as shown in Figure 1 . Risks at age 70 were 51% (95% confidence interval: 42–60%) for women in the highest quintile of PRS compared with 21% (14–31%) in the lowest. Similar plots for each of the two cohorts individually are presented in Supplementary Figure S1 online.
Analysis of PRS and family history
To explore the joint relationship of the PRS and family history regarding risk, we added the BOADICEA 10-year risk score to the Cox models and examined the effect of the PRS score adjusted for family history. For the set of individuals with these scores, the HR associated with the PRS in the combined data set was 1.36 (P = 2 × 10−6), whereas with the BOADICEA the 10-year score in the model the HR was only slightly reduced (1.34; P = 1 × 10−5). The BOADICEA 10-year risk estimate was also a significant predictor (HR = 1.1; P = 9 × 10−4) of BC risk. There was no evidence of an interaction between the PRS and BOADICEA 10-year risk (P = 0.31). Supplementary Figure S2 online shows the Kaplan–Meier plots for the lowest, middle, and highest tertiles of the baseline BOADICEA 10-year risk.
Figure 2 displays a plot of the BOADICEA lifetime risk plotted against the estimated remaining lifetime risk based on the BOADICEA/PRS score with indicators of the 20 and 25% risk categories, which would be considered cutoffs for recommending screening breast MRI. Table 3 shows the numbers of women in each of the risk quadrants for the two thresholds. For example, assuming the 20% threshold for MRI screening, 249 women out of 1,585 (16%) moved from below the threshold to above this threshold.
Scatter plot of BOADICEA lifetime risk against estimated lifetime risk based on the combination of BOADICEA score and the individual PRS. In the bottom panel, solid horizontal and vertical lines indicate the 20% threshold of lifetime risk, and the dashed lines denote the 25% threshold. Each red dot corresponds to an individual woman in the prospective cohort. Those in the upper left and lower right quadrants would be those who potentially could have a change in screening recommendations based on current guidelines. PRS, polygenic risk score.
Discussion
The results of this study show that using even a subset of the current ~96 breast cancer–associated SNPs can provide a potentially useful stratification of women into risk groups. However, the SNPs that we did not include in our study are, in general, rarer and/or have smaller effect size, so we believe we have captured a significant proportion of the known genetic variance of BC due to common alleles of small effect. Based on the theoretical standard deviation of the score calculated from 77 SNPs by Mavaddat et al.,5 we calculate that our PRS score captures approximately two out of three of the genetic variances represented in the more recent panel. It is likely that inclusion of more complete sets of SNPs would further increase the discriminatory power. To our knowledge, this is the first prospective study (familial or otherwise) to demonstrate the ability of such SNP panels to predict breast cancer outcome. Sawyer et al.7 estimated an HR of 2.08 for the lowest quartile compared with the highest quartile in assessing the risk of contralateral BC using a PRS based on 22 SNPs. However, this was a retrospective analysis in which women who presented with bilateral BC were compared with unilateral cases. This compares with the HR of 3.18 for the highest and lowest quintiles in our prospective analysis based on a PRS composed of 24 SNPs. Comparing familial BC cases to controls, the Sawyer study found an area under the curve (AUC) of 0.64 for predicting BC based on their PRS; in our prospective analysis, we found an AUC of 0.59 (95% confidence interval: 0.55–0.63). The absolute risks associated with women in the highest quintile of PRS were quite high, but it must be noted that these women in the BCFR were selected for genotyping based on having a family history and women/families enrolled in kConFab are selected on the basis of their family history.
Both the American Cancer Society8 and the National Comprehensive Cancer Network17 guidelines propose that women with a lifetime risk for BC above 20–25% should receive MRI screening. Using the BOADICEA algorithm to predict lifetime risk and assuming the 25% threshold, 14% of women in this familial cohort would theoretically have a change in management (i.e., screening or prevention recommendations); with the lower threshold of 20%, this figure increases to 23%. However, these estimates are based on the HRs for the PRS estimated from the data and thus would not be valid estimates of risk and are specific to the risk distribution in this set of selected families. However, this does demonstrate how the PRS can be used to more effectively target screening/prevention choices in BRCA1/2-negative women with a family history of BC.
In summary, we have shown that SNP panels can be a useful adjunct to genetic testing for high penetrance genes in women with a family history of BC. Inclusion of risk scores based on BC-associated SNPs in risk assessment can provide more accurate risk prediction than family history alone and can influence recommendations for cancer screening and prevention modalities for high-risk women.
Disclosure
P.K. is a consultant for Merck and has provided expert testimony for AstraZeneca. D.E.G. has received royalties from patents on the BRCA1 and BRCA2 genes from the University of Utah that are licensed to Myriad Genetics. G.M. has provided expert testimony and is a consultant for AstraZeneca. I.L.A. is an inventor on a patent held by Mt. Sinai Hospital in Toronto, Canada. A.M. is a stockholder in, and Chief Scientific Officer of, Covariance Biosciences, Inc. The other authors declare no conflict of interest.
References
National Institutes of Health. Precision Medicine Initiative Cohort Program, 2015. http://www.nih.gov/precisionmedicine/. Accessed 5 May 2015.
Michailidou K, Beesley J, Lindstrom S, et al.; BOCS; kConFab Investigators; AOCS Group; NBCS; GENICA Network. Genome-wide association analysis of more than 120,000 individuals identifies 15 new susceptibility loci for breast cancer. Nat Genet 2015;47:373–380.
Michailidou K, Hall P, Gonzalez-Neira A, et al.; Breast and Ovarian Cancer Susceptibility Collaboration; Hereditary Breast and Ovarian Cancer Research Group Netherlands (HEBON); kConFab Investigators; Australian Ovarian Cancer Study Group; GENICA (Gene Environment Interaction and Breast Cancer in Germany) Network. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat Genet 2013;45:353–361.
Pharoah PD, Antoniou AC, Easton DF, Ponder BA. Polygenes, risk prediction, and targeted prevention of breast cancer. N Engl J Med 2008;358:2796–2803.
Mavaddat N, Pharoah PD, Michailidou K, et al. Prediction of breast cancer risk based on profiling with common genetic variants. J Natl Cancer Inst 2015;107:.
Antoniou AC, Beesley J, McGuffog L, et al.; Ontario Cancer Genetics Network; SWE-BRCA; HEBON; EMBRACE; GEMO; GEMO; Breast Cancer Family Registry; kConFab; CIMBA. Common breast cancer susceptibility alleles and the risk of breast cancer for BRCA1 and BRCA2 mutation carriers: implications for risk prediction. Cancer Res 2010;70:9742–9754.
Sawyer S, Mitchell G, McKinley J, et al. A role for common genomic variants in the assessment of familial breast cancer. J Clin Oncol 2012;30:4330–4336.
Saslow D, Boetes C, Burke W, et al.; American Cancer Society Breast Cancer Advisory Group. American Cancer Society guidelines for breast screening with MRI as an adjunct to mammography. CA Cancer J Clin 2007;57:75–89.
National Collaborating Center for Cancer (UK). Familial Breast Cancer: Classification and Care of People at Risk of Familial Breast Cancer and Management of Breast Cancer and Related Risks in People with a Family History of Breast Cancer. National Collaborating Centre for Cancer (UK), 2013. http://www.ncbi.nlm.nih.gov/books/NBK247567/. Accessed 4 August 2015.
John EM, Hopper JL, Beck JC, et al.; Breast Cancer Family Registry. The Breast Cancer Family Registry: an infrastructure for cooperative multinational, interdisciplinary and translational studies of the genetic epidemiology of breast cancer. Breast Cancer Res 2004;6:R375–R389.
Terry MB, Phillips KA, Daly MB, et al. Cohort profile: the breast cancer prospective family study cohort (ProF-SC). Int J Epidemiol 2015; e-pub ahead of print 13 July 2015.
Osborne RH, Hopper JL, Kirk JA, Chenevix-Trench G, Thorne HJ, Sambrook JF. kConFab: a research resource of Australasian breast cancer families. Kathleen Cuningham Foundation Consortium for Research into Familial Breast Cancer. Med J Aust 2000;172:463–464.
Phillips KA, Butow PN, Stewart AE, et al.; kConFab Investigators. Predictors of participation in clinical and psychosocial follow-up of the kConFab breast cancer family cohort. Fam Cancer 2005;4:105–113.
Weeks DE, Ott J, Lathrop GM. SLINK: a general simulation program for linkage analysis. Am J Hum Genet 1990;47:A204.
Antoniou AC, Cunningham AP, Peto J, et al. The BOADICEA model of genetic susceptibility to breast and ovarian cancers: updates and extensions. Br J Cancer 2008;98:1457–1466.
MacInnis RJ, Bickerstaffe A, Apicella C, et al. Prospective validation of the breast cancer risk prediction model BOADICEA and a batch-mode version BOADICEA Centre. Br J Cancer 2013;109:1296–1301.
National Comprehensive Cancer Network. NCCN guidelines: genetic/familial high-risk assessment: breast and ovarian (version 2.2014). http://www.nccn.org/professionals/physician_gls/pdf/genetics_screening.pdf.Accessed 1 October 2015.
Acknowledgements
This research was supported by a grant from Cancer Australia’s Priority-driven Collaborative Cancer Research Scheme no. 566791 (to G.M.) and by NIH grant U19 CA 148065-01 (DRIVE, part of the GAME-ON initiative) and NIH grant R01CA155767 (to D.E.G.). The Breast Cancer Family Registry is supported by NIH grants R01 CA159868 and UM1 CA164920 from the National Cancer Institute.
The content of this article does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centers in the BCFR, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government or the BCFR. K.A.P. is an Australian National Breast Cancer Foundation Practitioner Fellow. I.L.A. holds the Anne and Max Tanenbaum Chair in Molecular Medicine at Mount Sinai Hospital and the University of Toronto.
We thank Eveline Niedermayr, all the kConFab research nurses and staff, the heads and staff of the Family Cancer Clinics, and the investigators and staff of the kConFab Clinical Follow-Up Study, which has received funding from the NHMRC, the National Breast Cancer Foundation, Cancer Australia, and the US National Institutes of Health, for their contributions to this resource. kConFab is supported by a grant from the National Breast Cancer Foundation and was previously supported by the National Health and Medical Research Council (NHMRC); the Queensland Cancer Fund; the Cancer Councils of New South Wales, Victoria, Tasmania, and South Australia; and the Cancer Foundation of Western Australia. Most importantly, the authors thank all the families enrolled in the BCFR and kConFab resources for their willingness to participate in research; without them, the work presented here would not be possible.
Author information
Authors and Affiliations
Consortia
Corresponding author
Supplementary information
Supplementary Information
(ZIP 793 kb)
Rights and permissions
About this article
Cite this article
Li, H., Feng, B., Miron, A. et al. Breast cancer risk prediction using a polygenic risk score in the familial setting: a prospective study from the Breast Cancer Family Registry and kConFab. Genet Med 19, 30–35 (2017). https://doi.org/10.1038/gim.2016.43
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gim.2016.43
Keywords
This article is cited by
-
Polygenic Risk Scores for Breast Cancer
Current Breast Cancer Reports (2024)
-
Clinical utility of polygenic risk scores: a critical 2023 appraisal
Journal of Community Genetics (2023)
-
Capturing additional genetic risk from family history for improved polygenic risk prediction
Communications Biology (2022)
-
The role of genomics in global cancer prevention
Nature Reviews Clinical Oncology (2021)
-
The Polygenic Score Catalog as an open database for reproducibility and systematic evaluation
Nature Genetics (2021)