Abstract
Purpose:
Glycogen storage disease (GSD) is an umbrella term for a group of genetic disorders that involve the abnormal metabolism of glycogen; to date, 23 types of GSD have been identified. The nonspecific clinical presentation of GSD and the lack of specific biomarkers mean that Sanger sequencing is now widely relied on for making a diagnosis. However, this gene-by-gene sequencing technique is both laborious and costly, which is a consequence of the number of genes to be sequenced and the large size of some genes.
Methods:
This work reports the use of massive parallel sequencing to diagnose patients at our laboratory in Spain using either a customized gene panel (targeted exome sequencing) or the Illumina Clinical-Exome TruSight One Gene Panel (clinical exome sequencing (CES)). Sequence variants were matched against biochemical and clinical hallmarks.
Results:
Pathogenic mutations were detected in 23 patients. Twenty-two mutations were recognized (mostly loss-of-function mutations), including 11 that were novel in GSD-associated genes. In addition, CES detected five patients with mutations in ALDOB, LIPA, NKX2-5, CPT2, or ANO5. Although these genes are not involved in GSD, they are associated with overlapping phenotypic characteristics such as hepatic, muscular, and cardiac dysfunction.
Conclusions:
These results show that next-generation sequencing, in combination with the detection of biochemical and clinical hallmarks, provides an accurate, high-throughput means of making genetic diagnoses of GSD and related diseases.
Genet Med 18 10, 1037–1043.
Similar content being viewed by others
Introduction
The term “glycogen storage disease” (GSD) refers to a group of disorders characterized by genetically determined errors of glycogen metabolism. Twenty-three types of GSD are currently recognized, covering a broad clinical spectrum involving different organs; however, the liver, skeletal muscle, heart, and sometimes the central nervous system are those most commonly affected. They are classified depending on the organ affected and the enzyme deficiency involved. GSD types I, III, 0, XI, IX, VI, and IV affect the liver (80% of hepatic GSD is type I, III, or IX 1,2), types II, IIIa, V, VII, IXd X, XII, XIII, and XIV affect the muscles, and type IIA, IIb, and PRKAG2 deficiency involve myopathy/cardiomyopathy. Some GSD types can affect both the liver and muscles (III and IXb).3
The overall incidence of GSD in the population is estimated at 1 case per 2,000–43,000.1 Liver-affecting GSD types involve hepatomegaly and hypoglycemia, with the consequence of poor glucose distribution throughout the body. Patients with muscle- and heart-affecting GSD experience exercise intolerance, often followed by notable rhabdomyolysis.3 Interestingly, phenotypic variation is wide, and the disease may take different clinical courses even though the same enzyme is involved. Variation is also seen in the age of onset of symptoms and morbidity and mortality; depending on the specific mutation involved, the prognosis may be favorable or unfavorable. Neonatal and infantile forms of GSD usually are more severe, whereas other forms are relatively asymptomatic or may cause only exercise intolerance.
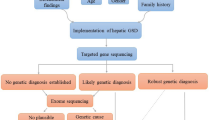
Early diagnosis is important if quality of life is to be improved and appropriate treatment is to be provided (when possible). Identifying the genetic background of patients with GSD may help in their counseling and that of their relatives. However, the accurate classification of GSD is no easy task. Mutation screening by conventional Sanger sequencing has been the gold standard in this respect for many years. However, this method can only examine one gene at a time, exon by exon. Some clinical laboratories still rely on even less reliable and time-consuming assays of glycogen-processing enzymes. For some forms of GSD, these assays can be performed using enzymes from fibroblasts, erythrocytes, or lymphocytes (type IIb, II, IIIa, IV, or VII), but for others, liver or muscle biopsies must be performed. Molecular methods therefore provide the best way of diagnosing and classifying GSD, but they need to be more rapid and cost-effective.
Fortunately, recent developments in high-throughput sequence capture have made next-generation sequencing feasible for use in routine genetic diagnosis.4 This very cost-effective technology is particularly appropriate for screening for mutations in disorders with highly heterogeneous genetic backgrounds, such as GSD, congenital disorder of glycosylation, lysosomal disorders, and mitochondrial disorders5,6,7,8 In addition, its ability to detect mutations in large genes and to identify copy number variations is very advantageous. The implementation of massive parallel sequencing has begun to revolutionize the field of genetic diagnosis. For example, for a large gene such as AGL, which has clear hallmarks, conventional genetic analysis involves the amplification of 34 exons plus the corresponding contamination controls and plus subsequent bidirectional Sanger sequencing. Massive parallel sequencing, in contrast, allows all exons to be sequenced at once, reducing costs and the time involved. Massive parallel sequencing technology generates large amounts of sequence data, and adding specific sequence tags (DNA bar codes) to each sample allows for pooled testing; this further reduces costs and time requirements, although, of course, pooling requires several patients be sequenced together with different disorders. The captured data are prioritized by matching them against patient clinical and biochemical hallmarks; without phenotype information, genome analysis would be of limited medical value.9
This article reports the genetic analysis of a cohort of 47 patients whose blood was sent to our laboratory for genetic diagnosis of suspected GSD. Massive parallel sequencing—via targeted exome sequencing (TES) or clinical exome sequencing (CES)—detected pathogenic mutations in 23 patients, 18 in previously described GSD-associated genes and 5 in the non-GSD-associated genes ALDOB, LIPA, CPT2, NKX2-5, and ANO5.
Materials and Methods
Blood samples from 47 patients suspected of having GSD were sent to the genetics laboratory at the Centro de Diagnóstico de Enfermedades Moleculares in Madrid, Spain, for genetic analysis. High-purity DNA was extracted from whole blood using the MagNA Pure Compact Kit (Roche Applied Biosciences) following the manufacturer’s protocol. DNA was quantified using picogreen (Invitrogen, Carlsbad, CA). Prior to analysis, written informed consent for genetic testing was obtained from all patients or their legal guardians.
Two massive parallel sequencing methods were used in an attempt to identify the mutations causing the suspected GSD: TES and CES. Sixteen patients were first examined by TES involving the use of a customized (Haloplex) panel (Agilent Technologies, Santa Clara, CA) of 111 genes involved in metabolic disorders, including 22 GSD-associated genes (AGL, ALDOA, ENO3, G6PC, GAA, GBE1, LDHA, GYG1, GYS1, GYS2, PFKM, PGAM2, PGM1, PHKA1, PHKA2, PHKB, PHKG1, PHKG2, PYGL, PYGM, SLC37A4, SLC2A2). The DNA samples were only examined for GSD genes or genes related to its pathological phenotypes. Incidental findings in genes unrelated to the clinical/biochemical phenotypes were ignored. A total of 346 GSD exons plus 50 bp of their flanking introns were captured. The minimum coverage achieved was 20× for 95% of the targeted bases; for AGL, GBE1, GYS2, and PHKA1, however, <20× coverage was achieved for 10% of the targeted bases. All the uncovered regions belonged to intronic sequences. The mean depth of coverage was 440× (range: 173–921×; Table 1 ). Each patient showed an average 1,200 sequence variants.
To improve the diagnostic yield, 43 patients (including 12 previously analyzed by TES plus 31 consecutive samples received for genetic diagnosis) were examined by CES using the Illumina Clinical-Exome Sequencing TruSight One Gene Panel. This panel includes all the known disease-associated genes described in the OMIM database until 2013, and captures 62,000 exons and their flanking intronic regions. A minimum coverage of 20× was achieved for 99% of the GSD targeted bases (mean depth of coverage of 83.6×). An average of 8,300 variants was identified per patient.
In both TES and CES, the libraries generated were sequenced using 250-bp paired-end reads using the Illumina MiSeq or Nextseq500 next-generation sequencing platforms. The Fastq files produced were examined using the DNAnexus platform (https://platform.dnanexus.com) to allow subsequent mapping and the generation of variant calling files. These variant calling files were analyzed using VariantStudio Data Analysis Software (Illumina, San Diego, CA). Synonymous variants and those with minor allele frequencies (>1% in dbSNP) were excluded. The remaining single-nucleotide variants were prioritized as follows: (i) variants in genes previously associated with the observed phenotype and that showed the expected pattern of inheritance; (ii) variants annotated in the Human Gene Mutation Database; (iii) loss-of-function mutations not previously reported (nonsense, splice site junction mutations, out-of-frame deletions or insertions); and (iv) candidate missense variants with pathogenicity scores as determined by SIFT (http://sift.jcvi.org/www/SIFT_enst_submit.html), Polyphen2 software (http://genetics.bwh.harvard.edu/pph2/), and MutationTaster (http://www.mutationtaster.org/). Genes with potentially pathogenic mutations were assessed in the context of the patient phenotype according to OMIM criteria (http://www.omim.org/).
Variants selected by these criteria were confirmed by conventional Sanger sequencing using the BigDye Terminator Cycle Sequencing kit (Applied Biosystems, Foster City, CA) using both the patients’ genomic DNA and that of their parents if available.
Results
When TES was used to determine the diagnosis rate (using a reduced number of genes to restrict incidental findings), three patients were detected bearing biallelic mutations in GAA (P1), AGL (P2), or PYGL (P18) ( Table 2 ). Another patient was detected with only a paternal pathogenic mutation in PHKB; she may have been simply a carrier of GSD or may have had a further undetected mutation in the maternal gene (P9). Because 111 genes related to metabolic disorders were captured by this gene panel, a patient (P19, Table 3 ) was detected carrying the most common mutation in the non-GSD-associated ALDOB gene (p.Ala150Pro). Thus, TES diagnosed five patients (diagnosis rate close to 31% (5/16)).
Because the diagnosis rate was low, CES was performed. Among the 43 patients examined, 18 were found to have pathogenic mutations (14 in GSD-associated genes and 4 in nonassociated GSD genes) ( Tables 2 and 3 ). The most common defects were in AGL (GSD III) and PHKA2 (GSD IX). The diagnosis rate was 43%.
Overall, 22 mutations were detected in the GSD-associated genes of 18 patients, 11 of which have never before been reported. These novel mutations include four frameshift variants in AGL (c.348_373del26, c.2711_2717del7ins13, c.2151delT, and c.4391_4392delAT) and three in PHKA2 (c.1404dupT, c.2387_2388delCT, and c.2753delG), one nonsense change (c.104T>G) in AGL, one splicing mutation (c.1423+1G>A) in AGL, one missense variant (p.Arg576Gln) in PYGL, and one deletion (c.2862_2864delCCT) in PHKA2 ( Table 2 ). All of these were confirmed by Sanger sequencing. Allelic segregation was analyzed using parental DNA samples.
The CES technique detected mutations in four other genes: LIPA, CPT2, ANO5, and NKX2-5. Although these are not GSD-associated genes, their mutation gave rise to phenotypic characteristics overlapping those of GSD ( Table 3 ). Patient P20 was a carrier of two described mutations in LIPA (the gene coding for lysosomal acid lipase (LAL) MIM 613497): c.894G>A and c.398delC.10,11 The first affects the splicing process and is frequently associated with cholesteryl ester storage disease, a mild form of LAL deficiency; the second is associated with a much more severe form known as Wolman disease. The defect was confirmed by enzyme assay in dried blood samples that showed significantly reduced LAL activity (enzyme activity <0.02 nmol/punch/hour; reference range: 0.37–2.30 nmol/punch/hour). P21 harbored (in homozygosis) a described missense mutation (p.Ser113Leu) in CPT2, P22 had the most common mutation in ANO5 (c.191dupA; also in homozygosis), and P23 bore (in homozygosis) a previously described mutation in NKX2-5 (p.Arg25Cys).
Overall, the diagnosis rate for massive parallel sequencing was 49% (23/47)
Discussion
The present work reports the first extensive mutation spectrum for GSD in Spain. Pathogenic biallelic or X-linked mutations were detected in 22 patients. In one patient, a pathogenic mutation was detected in the paternally derived gene only. This patient might be a carrier of a described mutation in PHKB or bear a second mutation in genomic regions not analyzed (i.e., the carrier of a deep intronic mutation, a regulatory mutation, or a mutation far away from PHKB). Mutations were detected in GAA, AGL, PHKB, PHKA2, SLC37A4, and PYGL. More than three-quarters of the present patients bore mutations in AGL or PHKA2 (39% in AGL and 39% in PHKA2). This is in contrast with that found in other studies1 in which GSD type IX (involving PHKA2) was the most common form of the disease. It is also in contrast to the results for patients collected by the Spanish GSD association (AAEEG; http://www.glucogenosis.org/), among whom the most common disease forms are GSD V (McArdle disorder) and GSD II (Pompe disease). These two types of GSD have specific clinical and biochemical hallmarks. In Spain, such patients’ samples are sent to dedicated clinical laboratories.
GSD type III, due to the defect caused in the glycogen debranching enzyme, was the most common disease type detected in the present cohort. In general, patients with GSD type III present clear biochemical and clinical hallmarks and, in fact, the patients described in Table 2 present clear biochemical and clinical hallmarks that invite the analysis of AGL. It is possible that in the majority of patients with these hallmarks, Sanger sequencing to confirm AGL involvement could be performed. However, massive parallel sequencing is cheaper when dealing with large genes; each sample may cost less than €450 to process (consumables), whereas bidirectional Sanger sequencing plus the necessary contamination controls cost approximately €20 per exon. Furthermore, massive parallel sequencing avoids allele dropout, and in many cases allows genomic rearrangements to be detected. Such rearrangements could then be fully characterized using whole genomic arrays. In some patients, the election to sequence AGL is not straightforward and several genes have to be sequenced before the affected gene and its pathogenic mutation are found. It has been reported that after Sanger sequencing, patients suspected of having either GSD type IV or GSD type Ia were confirmed to have GSD type III; suspicion of the former disease types probably arose because the patients had not yet developed the full spectrum of symptoms at the time of clinical assessment or presented with atypical clinical symptoms.8,12 Massive parallel sequencing offers a complete definition of the captured gene without the need for stepwise testing and having to choose which gene to sequence first. Thus, based on the results of the present study and those of previous reports,8,12 massive parallel sequencing should be performed to confirm what would appear to be very clear GSD types.
Of the 22 mutations detected (mostly loss-of-function mutations—small deletions/insertions, splicing, or nonsense mutations), 11 were novel. It is noteworthy that all the mutations detected in AGL were loss-of-function mutations. This type of mutation accounts for nearly 82% of those deposited for this gene in the Human Gene Mutation Database. 13,14,15 Missense mutations are scarce.13,16,17 No mutation was prevalent, but c.348_373del26 was present in three mutant alleles (21%). Loss-of-function mutations in PHKA2 made up nearly 71% of the total; only 50% of the mutations deposited for this gene in the Human Gene Mutation Database belong to this category. Even though the majority of mutations affected just two genes, simple mutation screening would not have provided an accurate diagnosis. The missense change detected in PYGL (p.Arg576Gln) was classified as probably damaging by SIFT, polyphen2, and Mutation Taster software analysis, because it affected conserved amino acids. It was not detected in 6,500 exomes (Exome Variant Server database) or 1,000 genomes (1000g Project database), also suggesting it should be classified as disease-causing. The in-frame deletion detected in PHKA2 (p.Leu955del) is also very likely pathogenic because it affects a highly conserved amino acid.
Massive parallel sequencing reduces the costs and turnaround time associated with the use of a single capture reagent because several samples’ exons and genes can be analyzed simultaneously.8 The capture of GSD candidate genes has been used in a validation cohort8 and was shown to be 100% sensitive and specific. However, it has not been used for making diagnoses in a discovery cohort as described in the present work. The TES method only returned a very low diagnosis rate, even though the depth of coverage was high. This is probably explained by the fact that other disorders mimic GSD. Therefore, even though analysis involving a customized capture panel of relevant genes can sometimes greatly shorten the time required to reach a diagnosis, a broader analysis involving other genes causing diseases with phenotypes overlapping that of GSD can be useful. The use of customized panels based on clinical features more than on biochemical findings might help improve the diagnosis rate. However, it should be remembered that the use of extended gene panels might increase the number of incidental findings, and that the use of liver or muscle GSD gene panels alone might lead to misdiagnoses.
Another way of increasing the diagnosis rate might be to use whole-exome sequencing. However, the base pair coverage provided by whole-exome sequencing is not uniform. A gene panel with fewer genes than used in whole-exome sequencing but with better base pair coverage—such as that used in the present CES technique—might be more recommended. The use of the present extended CES panel increased the diagnosis rate to 43%. In addition, mutations in the non-GSD-associated ALDOB, LIPA (in patients with liver dysfunction), CPT2, ANO5, and NKX2-5 (in patients with muscle and cardiac disease phenotypes) genes were detected. After identification of these genetic defects, clinical phenotyping was newly performed. Other authors report massive parallel sequencing to have changed a diagnosis of congenital disorder of glycosylation on the discovery of mutations in the GSD-associated gene PGM1 (MIM 171900).18 Similarly, patients clinically diagnosed with GSD have been found, by this form of analysis, to have mutations in the congenital disorder of glycosylation-associated gene PMM2.19 Whole-exome sequencing or whole-genome sequencing of the samples from patients for whom no diagnosis was reached would be recommended.
The present work returned unexpected findings for five patients. In two patients with liver dysfunction, one carried in homozygosity the most common mutation seen in ALDOB (MIM 612724) and the other carried mutations associated with LIPA. Three patients with the muscular or cardiac phenotype had mutations in CPT2, NKX2-5, or ANO5. All had an atypical presentation of the disease, and the overlapping clinical and biochemical phenotypes may have led to clinical misdiagnoses. For example, LAL deficiency was ruled out by the clinicians of the patient with two mutations in LIPA given the very slight dyslipidemia seen. Similarly, the absence of plasma acylcarnitines meant no carnitine defect was suspected in P21. In both cases, the presence of specific mutations, c.894G>A or p.Ala113Leu, were probably responsible for this atypical phenotype. Re-evaluation of the clinical findings in close collaboration with clinicians allowed accurate diagnoses to be made.
Patient P23, who had a congenital heart defect, had an unexpected mutation in homozygosis: the mutation p.Arg25Cys already described for NKX2-5 (MIM 600584). To the best of our knowledge, p.Arg25Cys has always been detected in heterozygosity in congenital heart defects. All the clinical cardiac hallmarks associated with this defect were present in this patient. Its presence in homozygosis may explain the increased severity of disease suffered and the patient’s premature death. The patient also had other clinical features described here for the first time ( Table 3 ). These are probably the result of other malfunctions of this transcriptional factor in processes other than fetal heart development.
A prompt and accurate diagnosis is important if treatment that can avoid irreversible damage is to be provided. Reaching a diagnosis, however, can be a difficult task when dealing with heterogeneous pathologies involving defects in multiple genes. Some of the diagnoses made in this work allowed new treatments to be prescribed. For example, the patient with LAL deficiency (P20), originally diagnosed as having a form of GSD but who is now known to have cholesteryl ester storage disease, has now been included in a clinical trial for LAL replacement (http://www.synageva.com/). Similarly, the patient with ALDOB deficiency (P19), once thought to have a form of GSD, has now been prescribed a fructose-free diet and has improved considerably. A correct genetic diagnosis is, of course, essential if proper genetic counseling is to be given, for a management plan to be designed, and for an outcome to be predicted.
In summary, the present work shows the usefulness of massive parallel sequencing in diagnosing GSD, and in differentiating it from diseases with overlapping phenotypes. It is cost-effective and time-efficient, and it could prevent patients from receiving the wrong treatment for years on end. When required, CES can be used to broaden the number of analyzable genes beyond that used in TES, allowing the detection of mutations in non-GSD-associated genes causing symptoms that might overlap with those of clinical GSD.
Disclosure
The authors declare no conflict of interest.
References
Ozen H. Glycogen storage diseases: new perspectives. World J Gastroenterol 2007;13:2541–2553.
Hicks J, Wartchow E, Mierau G. Glycogen storage diseases: a brief review and update on clinical features, genetic abnormalities, pathologic features, and treatment. Ultrastruct Pathol 2011;35:183–196.
Laforêt P, Weinstein DA, Smit, PA. The glycogen storage diseases and related disorders. In: Saudubray JM, Berghe G, Walter JH (eds). Inborn Metabolic Diseases: Diagnosis and Treatment, 5th edn. Springer: Berlin, Germany, 2012:115–139.
Ng SB, Buckingham KJ, Lee C, et al. Exome sequencing identifies the cause of a mendelian disorder. Nat Genet 2010;42:30–35.
Jones MA, Bhide S, Chin E, et al. Targeted polymerase chain reaction-based enrichment and next generation sequencing for diagnostic testing of congenital disorders of glycosylation. Genet Med 2011;13:921–932.
DaRe JT, Vasta V, Penn J, Tran NT, Hahn SH. Targeted exome sequencing for mitochondrial disorders reveals high genetic heterogeneity. BMC Med Genet 2013;14:118.
Fernández-Marmiesse A, Morey M, Pineda M, et al. Assessment of a targeted resequencing assay as a support tool in the diagnosis of lysosomal storage disorders. Orphanet J Rare Dis 2014;9:59.
Wang J, Cui H, Lee NC, et al. Clinical application of massively parallel sequencing in the molecular diagnosis of glycogen storage diseases of genetically heterogeneous origin. Genet Med 2013;15:106–114.
Frebourg T. The challenge for the next generation of medical geneticists. Hum Mutat 2014;35:909–911.
Klima H, Ullrich K, Aslanidis C, Fehringer P, Lackner KJ, Schmitz G. A splice junction mutation causes deletion of a 72-base exon from the mRNA for lysosomal acid lipase in a patient with cholesteryl ester storage disease. J Clin Invest 1993;92:2713–2718.
Bernstein DL, Hülkova H, Bialer MG, Desnick RJ. Cholesteryl ester storage disease: review of the findings in 135 reported patients with an underdiagnosed disease. J Hepatol 2013;58:1230–1243.
Roscher A, Patel J, Hewson S, et al. The natural history of glycogen storage disease types VI and IX: long-term outcome from the largest metabolic center in Canada. Mol Genet Metab 2014;113:171–176.
Goldstein JL, Austin SL, Boyette K, et al. Molecular analysis of the AGL gene: identification of 25 novel mutations and evidence of genetic heterogeneity in patients with glycogen storage disease type III. Genet Med 2010;12:424–430.
Lucchiari S, Pagliarani S, Salani S, et al. Hepatic and neuromuscular forms of glycogenosis type III: nine mutations in AGL. Hum Mutat 2006;27:600–601.
Shen JJ, Chen YT. Molecular characterization of glycogen storage disease type III. Curr Mol Med 2002;2:167–175.
Sentner CP, Vos YJ, Niezen-Koning KN, Mol B, Smit GP. Mutation analysis in glycogen storage disease type III patients in the Netherlands: novel genotype-phenotype relationships and five novel mutations in the AGL gene. JIMD Rep 2013;7:19–26.
Cheng A, Zhang M, Okubo M, Omichi K, Saltiel AR. Distinct mutations in the glycogen debranching enzyme found in glycogen storage disease type III lead to impairment in diverse cellular functions. Hum Mol Genet 2009;18:2045–2052.
Timal S, Hoischen A, Lehle L, et al. Gene identification in the congenital disorders of glycosylation type I by whole-exome sequencing. Hum Mol Genet 2012;21:4151–4161.
Choi R, Woo HI, Choe BH, et al. Application of whole exome sequencing to a rare inherited metabolic disease with neurological and gastrointestinal manifestations: a congenital disorder of glycosylation mimicking glycogen storage disease. Clin Chim Acta 2015;444:50–53.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Vega, A., Medrano, C., Navarrete, R. et al. Molecular diagnosis of glycogen storage disease and disorders with overlapping clinical symptoms by massive parallel sequencing. Genet Med 18, 1037–1043 (2016). https://doi.org/10.1038/gim.2015.217
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gim.2015.217
Keywords
This article is cited by
-
Diagnostic yield of clinical exome sequencing as a first-tier genetic test for the diagnosis of genetic disorders in pediatric patients: results from a referral center study
Human Genetics (2022)
-
Clinical and genetic spectrum of glycogen storage disease in Iranian population using targeted gene sequencing
Scientific Reports (2021)
-
Diagnosis of hepatic glycogen storage disease patients with overlapping clinical symptoms by massively parallel sequencing: a systematic review of literature
Orphanet Journal of Rare Diseases (2020)
-
Improving the diagnosis of cobalamin and related defects by genomic analysis, plus functional and structural assessment of novel variants
Orphanet Journal of Rare Diseases (2018)