Abstract
Purpose:
This study was designed to delineate genetic contributions, if any, to sporadic forms of mild to moderate sensorineural hearing loss (SNHL) not related to GJB2 mutations (DFNB1) in a pediatric population.
Methods:
We recruited 11 non-DFNB1 simplex cases of mild to moderate SNHL in children. We applied whole-exome sequencing to all 11 probands. We used a filtering strategy assuming that de novo variants of known autosomal dominant (AD) deafness genes, biallelic mutations in autosomal recessive (AR) genes, monoallelic mutations in X chromosome genes for males, and digenic inheritance could be associated. Candidate variants first were prioritized with allele frequency in public databases and confirmed by a phase or a segregation test in each family. Additional information from the literature or public databases was used to identify strong candidate variants.
Results:
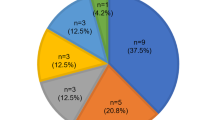
Strong candidate variants were detected in 5 of 11 probands (45.4%). A diverse mode of inheritance implicated the sporadic occurrence of the phenotype. AR mutations in OTOGL and SERPINB6 and digenic inheritance involving two deafness genes, GPR98 and PDZ7, were detected. A de novo AD mutation also was detected in TECTA and MYH14. No syndromic feature was detected in individuals with GPR98/PDZ7 or MYH14 variants in our cohort at this moment.
Conclusion:
Mild to moderate pediatric SNHL, even if sporadic, features a strong genetic etiology and can manifest via diverse modes of inheritance. In addition, a multidisciplinary approach should be used for a correct diagnosis.
Genet Med 17 11, 901–911.
Similar content being viewed by others
Introduction
Hereditary hearing loss contributes to more than 50% of hearing loss cases in pediatric population.1 Nonsyndromic hearing loss in the absence of other systemic manifestations accounts for ~70% of hereditary hearing loss. The most frequent mode of inheritance of nonsyndromic hearing loss is autosomal recessive (AR) (75–80% of cases).2,3,4 AR nonsyndromic hearing loss (OMIM 220700) typically has a congenital or prelingual onset.4 The degree of AR nonsyndromic hearing loss is usually severe or profound, although varying degrees of residual hearing can be observed in a subset of patients.5
Because of the huge impact of severe or profound hearing impairment in a pediatric population during speech and behavioral development, the main focus has been on etiological clarification in severe rather than milder hearing loss cases. However, mild to moderate hearing loss in early childhood, if not rehabilitated appropriately and in a timely manner, can also cause significant sequelae in school achievements and linguistic development. Early cochlear implantation is the only option for auditory rehabilitation of the majority of severe or total hearing loss. By contrast, diverse options are available for the management of mild to moderate hearing loss, depending on the degree and aggravation rate. Consequently, understanding the etiology of milder hearing loss can provide insight into the selection of appropriate auditory rehabilitation.
To date, mutations in 56 autosomal genes have been identified as a cause of AR nonsyndromic hearing loss, indicating extreme etiologic heterogeneity (http://hereditaryhearingloss.org/main.aspx?c=.HHH&n=86307). Most of the alterations in these AR genes are associated with severe hearing loss. However, a subset of deafness caused by GJB2 mutations (DFNB1), particularly in individuals carrying the p.V37I allele, is associated with mild to moderate hearing loss.6,7,8 Recessive mutations in the OTOG, OTOGL, and STRC genes also are implicated in moderate hearing loss, rather than severe to profound hearing loss, in children.9,10,11 Alterations in autosomal dominant (AD) deafness genes can manifest expectantly as hearing loss starting in childhood. Nevertheless, no extensive study has been conducted to delineate comprehensively the molecular genetic etiology of mild to moderate sensorineural hearing loss (SNHL), particularly a sporadic form, in a pediatric population. Lack of information regarding this issue in the literature encouraged us to expand our interests in genetic hearing loss further to milder hearing loss. Here we show diverse modes of inheritance involving sporadic moderate hearing loss in childhood.
Materials and Methods
Ethical considerations
This study of mild to moderate hearing loss in children was approved by the institutional review boards at the Seoul National University Hospital (IRBY-H-0905-041-281) and Seoul National University Bundang Hospital (IRB-B-1007-105-402). Informed consent was obtained from all participants in this study. For the child participants, written informed consent was obtained from the parents or guardians.
Study participants and audiometric evaluation
Clinical evaluation of all affected individuals by an ear, nose, and throat surgeon included medical and developmental history interviews, physical examinations, pure-tone audiometry, and imaging studies (high-resolution temporal bone computed tomography and/or magnetic resonance imaging) whenever possible. We referred all of our subjects to ophthalmologists and neuropediatricians to rule out any syndromic feature.
All subjects underwent pure-tone audiometry in a sound-proof room, with air-conduction and bone-conduction recordings. Air-conduction pure-tone average thresholds in the conversational frequencies of 0.5, 1, 2, and 4 kHz were measured for each ear and used to define deafness severity. Hearing level was labeled as subtle (16–25 dB), mild (26–40 dB), moderate (41–70 dB), severe (71–95 dB), or profound (95 dB). We regularly performed serial audiograms to detect any noticeable aggravation of hearing impairment after their participation in this study, and we provided timely auditory rehabilitation. The pure-tone average across these frequencies was calculated and reported as decibels of hearing loss. For SB106-198, on whom pure-tone audiometry could not be performed because he was too young, brainstem response threshold evaluations and/or auditory steady-state response(s) were recorded, and mean thresholds at frequencies in the 0.5- to 4-kHz range were averaged to approximate the pure-tone average. From September 2010 through June 2012, we consecutively recruited unrelated pediatric subjects who manifested mild to moderate SNHL during their first visit regardless of age at the onset of hearing loss and progression. Hearing loss in SB106-198 and SB112-204 was clearly documented as prelingual; however, clarifying whether the onset of hearing loss in the other subjects in our pediatric cohort as truly prelingual or postlingual was difficult (Supplementary Table S1 online). SB115-207 and SB114-206, who were >15 years old at their first visit, also were included in our cohort because they and their parents had clear recollection of their SNHL beginning during their first decade. Pediatric subjects with severe or profound hearing loss at their first visit were excluded from this study. Individuals with syndromic, unilateral hearing loss or with inner ear anomalies, such as an enlarged vestibular aqueduct or incomplete partition type III, also were excluded from this study (Supplementary Figure S1 online). For the remaining 22 patients, GJB2 sequencing was conducted, and six DFNB1 subjects were excluded. Finally, five multiplex families were excluded irrespective of the inheritance pattern of hearing loss. As a result, our cohort comprised 11 subjects who manifested a sporadic form of mild to moderate SNHL without any family history of SNHL ( Figure 1a,b ). Probands and any family members also were recruited for segregation of the variants that we detected. The parents of SB66-116 and SB125-217 refused to provide blood samples. Blood samples from only one parent of SB114 and SB115 were provided.
Audiologic phenotype and family pedigree of the 12 probands. (a) Pure-tone audiograms of the 11 probands are shown (auditory steady state response (ASSR) for SB106-198). Thresholds of the 11 probands all fall within the range of mild to moderate sensorineural hearing loss (SNHL). Audiograms of the right ear are in red, while those of the left ear are in blue. (b) Pedigrees show that SNHL in our cohort is sporadic. (c) SB109-201 carrying OTOGL mutations show stable SNHL, whereas hearing thresholds of SB114-206 carrying SERPINB6 mutations demonstrate significant progression. (d) The audiogram configuration of SB119-211 carrying a single heterozygous mutation of MYH14 shows a striking similarity to those of already published affected individuals carrying MYH14 mutations.
DNA preparation and whole-exome sequencing
Whole blood (10 ml) was obtained from all probands in the 11 families and, if possible, their siblings and parents for the segregation study. Genomic DNA was extracted from peripheral blood as described previously,8 and that from our 11 probands was subjected to whole-exome sequencing (WES). A SureSelect 50 Mb Hybridization and Capture kit was used for WES, and sequencing was performed using a HiSeq2000. Read length for paired-end reads was 100 bp.
Detection of single-nucleotide variants and insertion/deletion polymorphisms
Reads were aligned to the human genome reference sequence (hg19) using BWA version 0.7.512 with the “MEM” algorithm. We used SAMTOOLS version 0.1.18,13 GATK version 2.4-714 for sorting, indexing, and local realignment of SAM/BAM files. Picard version 1.93 (http://picard.sourceforge.net) was used to mark duplicates. We used the BaseRecalibrator from GATK (based on known single-nucleotide polymorphisms (SNPs) and insertions/deletions (indels) from dbSNP137, Mills, and 1000G gold standard indel b37 sites and 1000G phase I indel b37 sites for base recalibration. Targeted gene variants were called by the Unified Genotyper in GATK and also were recalibrated by GATK based on dbSNP137, Mills indels, HapMap, and Omni. Finally, we used ANNOVAR15 to annotate the variants.
Filtering process
We selected exonic and splicing variants, including nonsynonymous variants and small indels. We selected variants with allele frequencies <1% that were reported previously in NHLBI-ESP 6500, the 1000 Genomes Project, and our in-house Korean database consisting of exomes from 192 Korean individuals. We then filtered out the variants that were reported in dbSNP137, but we included those in the flagged SNP database maintained from the University of California, Santa Cruz.
To prevent loss of a potential true causative variant, we hypothesized that any of the following inheritance patterns could be associated with the development of a sporadic form of moderate hearing loss in children: (i) de novo mutations in known AD deafness genes, (ii) biallelic mutations in AR genes, (iii) monoallelic mutations in X chromosome genes for boys, and (iv) digenic inheritance. We labeled the variants possibly corresponding to at least one of these patterns as “inheritance pattern–matched variants.” In brief, these inheritance pattern–matched variants were chosen solely based on WES data from the probands. We classified the inheritance pattern–matched variants into groups according to the inheritance pattern in which the variants from the probands could cause deafness ( Figure 2 ).
Filtering strategies used to identify candidate variants. After annotation by ANNOVAR, rare variants are selected first with allele frequency <1%. If the variants are flagged single-nucleotide polymorphisms (SNPs) (from University of California, Santa Cruz) or not previously reported in dbSNP, they are subjected to the next filtering step. We check ethnicity-matched, normal-hearing control samples for the variants if the segregation of the variants in each family cannot be tested or is compatible with a known inheritance pattern of the gene. We designate the variants as “possible causal variants” if we cannot detect the variants from our control samples. Additional information from the literature or public databases is used to select strong candidate variants. VCF, variant call format.
If the inheritance pattern–matched variants cosegregated with the hearing loss phenotype in the segregation study, we then matched by ethnicity 138 normal-hearing control subjects (80 males and 58 females) as documented by pure-tone audiograms for the variants. When parental and sibling samples were missing because of their reluctance to join this study (i.e., SB125-217), we also subjected these samples to the control study to avoid overlooking any potential variants, even though the segregation study was not performed. The age of our normal-hearing controls ranged from 32 to 48 years old. To prevent loss of potential true causative variants, we applied an exclusion-based filtering strategy when we selected “possible candidate after segregation test” ( Table 1 ). Specifically, we excluded variants if our segregation test among family members revealed apparent discordance or if the detection frequency of the variant in our control chromosomes was more than 1/200.
Finally, we selected “strong candidate variants” among the possible causal variants in the case that the variants resided in the already-known deafness genes, the public database strongly supported their pathogenicity, or both. To assess the pathogenicity of possible candidates, we calculated a pathogenicity score16 by summing the SIFT (http://sift.jcvi.org/), PolyPhen2 (http://genetics.bwh.harvard.edu/pph2/), likelihood ratio test (http://www.molecularevolution.org/resources/lrt), MutationTaster (http://www.mutationtaster.org/), and GERP++ (http://mendel.stanford.edu/SidowLab/downloads/gerp/) scores to predict deleterious variants. The variants were classified as strong candidate variants if pathogenicity scores of two recessive variants were both equal to six.
Copy-number variant analysis
We adopted a read depth strategy to analyze copy-number variants (CNVs) using high-throughput sequencing data. Excavator version 2.216 was used to call CNVs to minimize any systematic biases, such as guanine-cytosine (GC) content, mappability, and exon length.16
Results
We analyzed the whole exomes of 11 unrelated pediatric subjects manifesting a sporadic form of mild to moderate SNHL. On average, we achieved 100× mean exome coverage, with 90% covered at >10×. Over 90% of unique reads were aligned with a mapping quality score >0, as compared with the reference (Supplementary Table S2 online).
After removing common variants with allele frequency >1% in public and in-house databases, we significantly reduced the number of candidates by including only inheritance pattern–matched variants among the novel SNPs in patients. By verifying the phase or segregation of the variants in normal-hearing parents and/or siblings of each family, we were able to reduce the number of possible candidates to seven or fewer in all 11 cases ( Table 1 , Supplementary Tables S3 and S4 online). At least one or more variants remained possible candidates for all 11 cases ( Table 1 , Supplementary Tables S3 and S4 online).
We found a strong candidate variant in 5 of the 11 probands (41.6%) ( Table 2 and Supplementary Table S3 online) after applying our filtering strategy. The strong candidate genes that caused mild to moderate SNHL in five probands were TECTA, OTOGL, GPR98/PDZD7, SERPINB6, and MYH14 ( Tables 1 and 2 , Figure 3 ). In detail, a known pathogenic heterozygous splice site variant of TECTA, c.5383+5delGTGA, was found in patient SB106-198. This de novo splice site variant was predicted to significantly affect canonical splicing, as predicted by the exonic splicing enhancers (ESE) finder (prediction score: 8.39 → 4.131) and also by the Berkeley Drosophila Genome Project (prediction score: 0.87 → 0.00) (http://www.fruitfly.org/seq_tools/splice.html). Paternity was confirmed at the molecular level to confirm that this missense variant of SB106-198 acted as a de novo allele. SB109-201, manifesting stable hearing thresholds over 2 years ( Figure 1c ), turned out to carry two definitely pathogenic mutations of OTOGL that lead to truncation of the protein. A novel splicing variant, GPR98 c.3022+2T>G, from patient SB112-204 was most likely pathogenic because this mutation would abolish canonical splicing as predicted by the ESE finder (prediction score: 2.9 → 0) (http://rulai.cshl.edu/cgi-bin/tools/ESE3/esefinder.cgi?Process=home). However, patient SB112-204 did not present any symptoms of nyctalopia, and ophthalmologic examinations, including electroretinography and optical coherence tomography, did not reveal any functional or structural retinal abnormalities (Supplementary Figure S2 online). Similarly, a novel SERPINB6 splicing variant (c.730-1G>A) detected in SB114-206 was predicted to be deleterious for a canonical splicing site (prediction score: 10.8 → −3.18 by the ESE finder). Also, a de novo MYH14 missense substitution, p.D529N, in SB119-211 was predicted to be pathogenic by Polyphen2 and SIFT and was evolutionarily significantly conserved among species (GERP++ score of 4.37). Paternity also was confirmed at the molecular level to ensure that this missense variant in SB119-211 acted as a de novo allele. Electromyography did not reveal any abnormalities indicative of peripheral neuropathy, thus suggesting that SNHL in SB119-211 was nonsyndromic.
Diverse mode of inheritance of moderate sensorineural hearing loss (SNHL) and mutations of several genes. (a) SB112-204 with moderate SNHL carries one mutant allele of PDZD7 descending from the father and the other mutant allele of GPR98 descending from the mother, manifesting SNHL in a digenic manner. SB119-211 and SB106-198, with moderate SNHL, show de novo occurrence of an MYH14 variant and a TECTA variant, respectively. An MYH14 variant affects the residues that are well conserved among species. (b) SB109-201 and SB114-206 segregate compound heterozygous mutations of OTOGL and SERPINB6 (please see Sanger traces), respectively, manifesting SNHL in an autosomal recessive manner.
In addition to single-nucleotide variants in the 11 samples, we also analyzed possible CNVs. We specifically focused on CNVs in known deafness genes and calculated the depth of coverage for the targeted regions (Supplementary Table S5 online). Indeed, there were several exons whose coverage was below 10×, in particular, exon 1 of some genes (presumably high GC content). It could be possible that some pathogenic CNVs may have resided in this area. However, we confirmed that most exons of the known deafness genes were sufficiently covered overall. Then we investigated CNVs using Excavator: (i) any deletion or duplication of alleles in an AR pattern or (ii) one AD allele (de novo). However, there were no deletions or duplications in the targeted regions.
Mild to moderate SNHL occurred in a Korean pediatric population via at least three types of inheritance modes: (i) biallelic mutations of AR genes (SB109-201, SB114-206), (ii) de novo occurrence of an AD mutation (SB119-211, SB106-198), and (iii) digenic inheritance (SB112-204). Depending on the causative gene, hearing loss progression greatly differed. Specifically, subject SB114-206, carrying SERPINB6 mutations, manifested significant aggravation of hearing thresholds over 2 years, whereas subject SB109-201, with OTOGL mutations, had stable hearing loss over 2 years ( Figure 1c ).
Discussion
A homozygosity mapping strategy using large consanguineous families has been used to discover prevalent and predominant deafness genes. However, the molecular etiology of deafness caused by low-prevalence genes in a sporadic form requires a completely different methodology. The prevalence of genetic disorders in pediatric subjects manifesting sporadic mild to moderate SNHL has not been reported accurately. To this end, we established a disease cohort comprising mainly pediatric cases of sporadic mild to moderated SNHL unrelated to GJB2 mutations. We subjected all samples from our 11 probands to WES, instead of using commercially available deafness panels, to address more effectively the proportion of genetic contribution to the patients’ milder SNHL. In this study we proposed a filtering strategy customized for the genetic diagnosis of milder SNHL through WES in small, nonconsanguineous families ( Figure 2 ).
Interestingly, we showed that at least three types of inheritance modes are implicated in the sporadic form of mild to moderate SNHL in a Korean pediatric population. These inheritance modes include (i) biallelic mutations of AR genes (OTOGL, SERPINB6), (ii) de novo occurrence of an AD mutation (MYH14, TECTA), and (iii) digenic inheritance (GPR98/PDZD7). This is in sharp contrast to the fact that AR inheritance accounts for the majority of prelingual severe to profound pediatric SNHL cases. The diverse inheritance modes of mild pediatric SNHL documented in our study indicate that a more sophisticated analytical approach is required after basic filtering of WES data from probands.
OTOGL at the DFNB84 locus and SERPINB6 at the DFNB91 locus reportedly cause mild to moderate SNHL in a pediatric population if mutated.11,17 OTOGL is expressed in Claudius cells, Hensen’s cells, outer hair cells, and also prominently in the three acellular gelatinous membranes (i.e., the cupula, the otoconical membrane, and the tectorial membrane). OTOGL is transcribed at high levels during the embryonic stage, after which its transcription decreases in the early postnatal stage and further so in the adult stages.11 Indeed, knocking down the otogelin-like protein encoded by OTOGL led to hearing loss and anatomic changes in the inner ear of zebrafish.11 Downregulation of this gene during the postnatal and adult stages supports this protein’s role mainly in anatomical structure development; it is likely not significantly associated with maintenance of the structures.11 Application of WES to delineate the etiology of bilateral mild to moderate SNHL in a French family recently discovered a biallelic nonsense mutation of OTOGL, and no audiogram hearing thresholds exceeded 70 dB. In line with the expression profile that supports a developmental rather than maintenance role of this protein, audiograms were reportedly stable during the 5-year follow-up period.18,19 Subject SB109-201 in our cohort also had stable hearing thresholds over 2 years ( Figure 1c ). By contrast, hearing loss caused by mutations in SERPINB6 (DFNB91) was strikingly different from that caused by OTOGL mutations in many aspects. To minimize nonspecific intracellular damage, the activities of serine and cysteine proteases are tightly regulated by inhibitors, such as those in the serpin superfamily. Among these, clade B serpins encoded by SERPINB6 are the predominant intracellular protease inhibitors. Absence of SERPINB6A in mice demonstrated age-related progressive hearing loss that was exaggerated at high frequencies.20 SERPINB6 mutations were recently reported to cause late–onset, postlingual, moderate SNHL.17,21 Alterations in this gene do not seem to contribute to congenital, prelingual, severe to profound SNHL based on the absence of mutations in this gene among a large cohort comprising such cases.17 The progressive nature of SNHL, as related to SERPINB6, was proposed even though its progression has not been validated by audiograms in the literature.17,21 Indeed, subject SB114-206, who carried SERPINB6 mutations in this study, showed significant deterioration of the hearing threshold over 2 years, which was in marked contrast to SB109-201, who possessed OTOGL mutations ( Figure 1c ). The age at onset of DFNB91-induced SNHL in humans has not been determined previously, but our study clearly indicates that SNHL onset occurred during the early teens, suggesting DFNB91 as a possible etiology of pediatric SNHL. Abrupt down-sloping, ski slope–type hearing loss, as occurred in subject SB114-206, caught our attention ( Figure 1a ) because alterations in TMPRSS3, a serine protease, are also known to cause progressive ski slope–type hearing loss.22,23 The fact that alterations in both protease and protease inhibitors cause ski slope–type SNHL may imply that high-frequency hearing is more vulnerable to imbalance between the activities of proteases and protease inhibitors.
In this study we also investigated the digenic inheritance of moderate SNHL involving two Usher syndrome type II (USH2)–related genes, GPR98 and PDZD7. In relation to USH2, SNHL is usually manifested to a moderate degree, although mild and severe degrees have also been reported.24 In our subject SB112-204, we identified a heterozygous mutant allele of GPR98, a known USH2 gene, along with a heterozygous PDZD7 mutant allele. PDZD7 was recently suggested to be a modifier of USH2. Specifically, a heterozygous mutant allele of PDZD7 exacerbates the course of retinitis pigmentosa in subjects carrying a heterozygous truncation mutation of USH2A or GPR98 in trans with the heterozygous mutant allele of PDZD7.25 Therefore, it was proposed that a subset of Usher syndrome may not be a Mendelian but a digenic or oligogenic disease.25 This suggestion is compatible with later observations that PDZD7 colocalized with usherin, whirlin, and GPR98 to the ankle-link region,25 as well as with observations that cytosolic domains of usherin and GPR98 can bind to PDZD7.26 Based on these observations, the role of PDZD7 was predicted to be involved in ankle-link complex scaffolding components.26 SB112-204 showed no retinal functional or structural abnormalities, however, and there was no evidence supporting the current existence of retinitis pigmentosa. The inability to detect any ophthalmologic abnormality in SB112-204 was not surprising considering the mean age of retinitis pigmentosa diagnosis in USH2. The median age of SNHL diagnosis in USH2 was reportedly 5 years (8 months to 31 years), whereas that of Usher syndrome diagnosis was 34.5 years (8–76 years).24 Mild to moderate SNHL may act as an important early biological marker for diseases that develop later.
SB119-211 had a de novo MYH14 mutation. This mutated allele was altered in a residue that was well conserved evolutionarily ( Figure 2a ). MYH14 encodes one of the heavy chains of the class II nonmuscle myosins (i.e., heavy chain 14). This gene resides within the DFNA4 locus,27,28 suggesting the possible contribution of this gene to deafness, even though the heterogeneity of this locus was reported subsequently.29,30 By evaluating four families with cosegregating moderate SNHL and a MYH14 mutation, Donaudy et al.31 reported a substantial contribution of MYH14 mutations to hereditary SNHL. Interestingly, the MYH14 mutation occurred as a de novo alteration in one of the four families. The phenotypic spectrum of this gene was recently expanded significantly by the discovery of a MYH14 variant responsible for peripheral neuropathy, myopathy, and hoarseness, in addition to mild to moderate SNHL.32 The cochlear expression pattern of this protein (i.e., higher expression in adults than during development) suggests that hearing loss may progress over time, which also was noted in the DFNA4 subjects with MYH14 mutations.31 However, detailed audiograms related to MYH14 mutations were not provided in previous studies. In this study we obtained an audiogram configuration for subject SB119-211 that was very similar to those of previously reported syndromic patients from the FC317 family who express a MYH14 mutation ( Figure 1d ),32 supporting a causal relationship between MYH14 p.D529N and SB119-211 SNHL.
The heterozygous splice-site variant of TECTA (c.5383+5delGTGA) seen in SB106-198 was detected initially in a UK family by segregating AD SNHL.33 It was recently reported to generate aberrant splicing products by exon 16 skipping, but not by reading frame disruption, in lymphoblastoid cell lines from affected subjects in a Brazilian family.34 Mutations in the TECTA gene result in either AD (DFNA8/12) or AR (DFNB21) SNHL.35,36 This variant affects the zonadhesion-zona (ZA-ZP) interdomain, and a previous study described nonprogressive, slightly down-sloping or flat-type, moderate to severe hearing loss, which is compatible with the audiogram configuration observed in patient SB106-198. More importantly, our study clearly shows that the onset of SNHL related to this splice-site variant was prelingual and also reports the de novo occurrence of an AD TECTA variant.
Using WES and a subsequent rigorous filtering strategy, we obtained a convincing molecular genetic diagnosis in at least 5 of the 11 pediatric patients (45.4%). Based on this result, more than 45.4% of mild to moderate SNHL unrelated to DFNB1 in a Korean pediatric population can be attributed to genetic alterations. The proportion could possibly be even higher than 45.4% because we could not finalize a causative variant for SNHL in a subset of our cohort because of the small family size. It is also possible that causative variants may be regulatory noncoding variants that were not detectable by exome sequencing. In addition, the sample size is too small to extrapolate the diagnostic yield of exome sequencing for hearing loss, and a larger-scale study is needed to draw a more solid conclusion. Nevertheless, this detection rate seems sufficiently high for further clinical efforts to identify a molecular genetic etiology in these patients. In this study we were able to attribute a strong genetic contribution to a sporadic form of mild to moderate SNHL unrelated to DFNB1 in a pediatric population. Diverse modes of inheritance were observed, and this should be considered when interpreting and filtering the results of molecular genetic testing in children with moderate SNHL and when counseling their families. In particular, a multidisciplinary approach including neurological and ophthalmological evaluations should be used to determine a correct diagnosis and to comprehensively manage these patients. Predicting the degree of progression based on molecular etiology would enable us to choose appropriate auditory rehabilitation.
Disclosure
The authors declare no conflict of interest.
References
Yiin RS, Tang PH, Tan TY . Review of congenital inner ear abnormalities on CT temporal bone. Br J Radiol 2011;84:859–863.
Denoyelle F, Weil D, Maw MA, et al. Prelingual deafness: high prevalence of a 30delG mutation in the connexin 26 gene. Hum Mol Genet 1997;6:2173–2177.
Shearer AE, Smith RJ . Genetics: advances in genetic testing for deafness. Curr Opin Pediatr 2012;24:679–686.
Morton CC, Nance WE . Newborn hearing screening–a silent revolution. N Engl J Med 2006;354:2151–2164.
Smith RJ, Hone S . Genetic screening for deafness. Pediatr Clin North Am 2003;50:315–329.
Chan DK, Schrijver I, Chang KW . Connexin-26-associated deafness: phenotypic variability and progression of hearing loss. Genet Med 2010;12:174–181.
Kobayashi K, Oguchi T, Asamura K, et al. Genetic features, clinical phenotypes, and prevalence of sensorineural hearing loss associated with the 961delT mitochondrial mutation. Auris Nasus Larynx 2005;32:119–124.
Kim SY, Park G, Han KH, et al. Prevalence of p.V37I variant of GJB2 in mild or moderate hearing loss in a pediatric population and the interpretation of its pathogenicity. PLoS One 2013;8:e61592.
Francey LJ, Conlin LK, Kadesch HE, et al. Genome-wide SNP genotyping identifies the Stereocilin (STRC) gene as a major contributor to pediatric bilateral sensorineural hearing impairment. Am J Med Genet A 2012;158A:298–308.
Schraders M, Ruiz-Palmero L, Kalay E, et al. Mutations of the gene encoding otogelin are a cause of autosomal-recessive nonsyndromic moderate hearing impairment. Am J Hum Genet 2012;91:883–889.
Yariz KO, Duman D, Seco CZ, et al. Mutations in OTOGL, encoding the inner ear protein otogelin-like, cause moderate sensorineural hearing loss. Am J Hum Genet 2012;91:872–882.
Li H, Durbin R . Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010;26:589–595.
Li H, Handsaker B, Wysoker A, et al.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009;25:2078–2079.
McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297–1303.
Wang K, Li M, Hakonarson H . ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010;38:e164.
Magi A, Tattini L, Cifola I, et al. EXCAVATOR: detecting copy number variants from whole-exome sequencing data. Genome Biol 2013;14:R120.
Cengiz FB, Duman D, Sirmaci A, et al. Recurrent and private MYO15A mutations are associated with deafness in the Turkish population. Genet Test Mol Biomarkers 2010;14:543–550.
Askes SH, Bahreman A, Bonnet S . Activation of a photodissociative ruthenium complex by triplet-triplet annihilation upconversion in liposomes. Angew Chem Int Ed Engl 2014;53:1029–1033.
Bonnet C, Louha M, Loundon N, et al. Biallelic nonsense mutations in the otogelin-like gene (OTOGL) in a child affected by mild to moderate hearing impairment. Gene 2013;527:537–540.
Tan J, Prakash MD, Kaiserman D, Bird PI . Absence of SERPINB6A causes sensorineural hearing loss with multiple histopathologies in the mouse inner ear. Am J Pathol 2013;183:49–59.
Sirmaci A, Erbek S, Price J, et al. A truncating mutation in SERPINB6 is associated with autosomal-recessive nonsyndromic sensorineural hearing loss. Am J Hum Genet 2010;86:797–804.
Chung J, Park SM, Chang SO, et al. A novel mutation of TMPRSS3 related to milder auditory phenotype in Korean postlingual deafness: a possible future implication for a personalized auditory rehabilitation. J Mol Med 2014;92:651–663.
Weegerink NJ, Schraders M, Oostrik J, et al. Genotype-phenotype correlation in DFNB8/10 families with TMPRSS3 mutations. J Assoc Res Otolaryngol 2011;12:753–766.
Abadie C, Blanchet C, Baux D, et al. Audiological findings in 100 USH2 patients. Clin Genet 2012;82:433–438.
Ebermann I, Phillips JB, Liebau MC, et al. PDZD7 is a modifier of retinal disease and a contributor to digenic Usher syndrome. J Clin Invest 2010;120:1812–1823.
Grati M, Shin JB, Weston MD, et al. Localization of PDZD7 to the stereocilia ankle-link associates this scaffolding protein with the Usher syndrome protein network. J Neurosci 2012;32:14288–14293.
Blum K, Sheridan PJ, Wood RC, Braverman ER, Chen TJ, Comings DE . Dopamine D2 receptor gene variants: association and linkage studies in impulsive-addictive-compulsive behaviour. Pharmacogenetics 1995;5:121–141.
Mirghomizadeh F, Bardtke B, Devoto M, et al. Second family with hearing impairment linked to 19q13 and refined DFNA4 localisation. Eur J Hum Genet 2002;10:95–99.
Yang T, Pfister M, Blin N, Zenner HP, Pusch CM, Smith RJ . Genetic heterogeneity of deafness phenotypes linked to DFNA4. Am J Med Genet A 2005;139:9–12.
Zheng J, Miller KK, Yang T, et al. Carcinoembryonic antigen-related cell adhesion molecule 16 interacts with alpha-tectorin and is mutated in autosomal dominant hearing loss (DFNA4). Proc Natl Acad Sci USA 2011;108:4218–4223.
Donaudy F, Snoeckx R, Pfister M, et al. Nonmuscle myosin heavy-chain gene MYH14 is expressed in cochlea and mutated in patients affected by autosomal dominant hearing impairment (DFNA4). Am J Hum Genet 2004;74:770–776.
Choi BO, Kang SH, Hyun YS, et al. A complex phenotype of peripheral neuropathy, myopathy, hoarseness, and hearing loss is linked to an autosomal dominant mutation in MYH14. Hum Mutat 2011;32:669–677.
Hildebrand MS, Morín M, Meyer NC, et al. DFNA8/12 caused by TECTA mutations is the most identified subtype of nonsyndromic autosomal dominant hearing loss. Hum Mutat 2011;32:825–834.
Lezirovitz K, Batissoco AC, Lima FT, et al. Aberrant transcript produced by a splice donor site deletion in the TECTA gene is associated with autosomal dominant deafness in a Brazilian family. Gene 2012;511:280–284.
Verhoeven K, Van Camp G, Govaerts PJ, et al. A gene for autosomal dominant nonsyndromic hearing loss (DFNA12) maps to chromosome 11q22-24. Am J Hum Genet 1997;60:1168–1173.
Mustapha M, Weil D, Chardenoux S, et al. An alpha-tectorin gene defect causes a newly identified autosomal recessive form of sensorineural pre-lingual non-syndromic deafness, DFNB21. Hum Mol Genet 1999;8:409–412.
Acknowledgements
This work is supported by the Korea Healthcare Technology R&D Project, Ministry for Health Welfare and Family Affairs, Republic of Korea (Grant A080588, HI11C133100000 (previously A111377) to B.Y.C., grant HT12C0014020014 to W.-Y.P.), and by the Seoul National University Bundang Hospital Research Fund (grant 04-2010-003 to B.Y.C.).
Author information
Authors and Affiliations
Corresponding authors
Supplementary information
Supplementary Figures and Tables
(ZIP 688 kb)
Rights and permissions
About this article
Cite this article
Kim, N., Kim, A., Park, K. et al. Whole-exome sequencing reveals diverse modes of inheritance in sporadic mild to moderate sensorineural hearing loss in a pediatric population. Genet Med 17, 901–911 (2015). https://doi.org/10.1038/gim.2014.213
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gim.2014.213
Keywords
This article is cited by
-
Novel homozygous variant in the PDZD7 gene in a family with nonsyndromic sensorineural hearing loss
BMC Medical Genomics (2022)
-
Full etiologic spectrum of pediatric severe to profound hearing loss of consecutive 119 cases
Scientific Reports (2022)
-
Molecular aetiology of ski-slope hearing loss and audiological course of cochlear implantees
European Archives of Oto-Rhino-Laryngology (2022)
-
A novel recessive PDZD7 bi-allelic mutation in an Iranian family with non-syndromic hearing loss
BMC Medical Genomics (2021)
-
Spectrum of genetic variants in moderate to severe sporadic hearing loss in Pakistan
Scientific Reports (2020)