Abstract
Purpose:
We show that a novel fragile X–related epigenetic element 2 FMR1 methylation test can be used along with a test for sex-determining region Y (SRY) to provide the option of combined fragile X syndrome and sex chromosome aneuploidy newborn screening.
Methods:
Fragile X–related epigenetic element 2, SRY, and FMR1 CGG repeat analyses were performed on blood and saliva DNA, and in adult and newborn blood spots. The cohort consisted of 159 controls (CGG <40), 187 premutation (CGG 56–170), and 242 full-mutation (CGG ~200–2,000) males and females, 106 sex chromosome aneuploidy individuals, and 151 cytogenetically normal controls.
Results:
At the 0.435 threshold, fragile X–related epigenetic element 2 analysis in males was robust on both blood DNA and newborn blood spots, with specificity and sensitivity of ~100% for full-mutation genotype. In females, the specificity was 99%, whereas half of full-mutation females were above the 0.435 threshold in both blood DNA and newborn blood spots. Furthermore, at this threshold, the test could not differentiate individuals with Klinefelter syndrome from female controls without using the SRY marker. When combined with SRY analysis, the test was consistent with most results for sex chromosome aneuploidies from karyotyping.
Conclusion:
Setting specific thresholds for fragile X–related epigenetic element 2 analysis and including the SRY marker provides the option to either include or exclude detection of sex chromosome aneuploidies as part of fragile X syndrome newborn screening.
Genet Med 2013:15(4):290–298
Similar content being viewed by others
Introduction
There are beneficial early therapy interventions for individuals clinically affected with sex chromosome aneuploidy or fragile X syndrome (FXS). Thus, a case can be made for including testing for these conditions in newborn screening programs.1 Sex chromosome aneuploidies have high frequencies in the general population. Klinefelter syndrome is found in up to 1 in 450–600 males,2,3 45,X Turner syndrome in 1 in 2,000 females,4 and 47,XXX in 1 in 1,000 females.4 Although some 47,XXX individuals display marked difficulties in speech and language, motor skills, and educational achievement,5 most of the individuals with 47,XXX do not manifest major medical problems.6 In contrast, girls with Turner syndrome suffer from a number of major medical problems including cardiac anomalies, short stature, and gonadal dysgenesis, which if detected early enough can be treated through cardiac intervention, growth hormone, or sex hormone therapy at puberty.7 Most men with Klinefelter syndrome have primary gonadal failure and benefit from testosterone replacement therapy from puberty. In addition, they and 47,XYY individuals have been reported to be at increased risk for autism spectrum disorder,8 and may therefore benefit from educational and behavioral interventions before puberty.2,9 This is pertinent for Klinefelter syndrome given that up to 70% of cases currently remain undiagnosed, and most of these diagnoses occur well into adulthood.2
We have developed a high-throughput FXS test targeting methylation of specific biomarker CpG sites within the FMR1 intron 1 region named fragile X–related epigenetic element 2 (FREE2). The test uses matrix-assisted laser desorption/ionization time-of-flight mass spectrometry system that can provide automated methylation analysis of thousands of DNA samples daily, at a lower cost as compared with standard FXS testing.10,11,12,13 This test also has the potential to be used for detection of sex chromosome aneuploidy.
FXS is the most common inherited form of intellectual disability, with a frequency of 1 in 3,600 in the general population.14 It is also one of the most common inherited conditions comorbid with autistic behaviors, and accounts for ~2–6% of all individuals diagnosed with autism (reviewed in ref. 15) FXS is usually caused by expansion of a trinucleotide CGG repeat within FMR1 to >200 repeats, termed full mutation (FM).16 In most cases, this causes silencing of FMR1 due to methylation of its promoter and loss of its protein product (fragile X mental retardation protein), which is essential for normal neurodevelopment.17,18,19 The more common small-expansion alleles, termed premutation (PM: 55–199 CGG repeats in 1 in ~300 individuals),20 and gray-zone alleles (45–54 CGG repeats 1 in ~ 30 individuals)21 do not cause the epigenetic silencing of FMR1 gene associated with FXS, but have been linked to late-onset neurological and reproductive disorders through an RNA toxicity mechanism.22,23
The FREE2 blood test identifies cognitively impaired FM males and females with specificity and sensitivity approaching 100%, but cannot distinguish between small-expansion carriers and healthy controls or high-functioning males with unmethylated FM alleles.11,12,13 This is highly advantageous for newborn, infant, or early childhood population screening because the test potentially detects only those who would directly benefit from early diagnosis.24
In this study, we performed technical validation for the FREE2 analysis in detection of FXS alleles in archival newborn and adult dried blood spots and venous blood DNA. We also explored the potential of the test to detect all sex chromosome aneuploidies with a view to expanding the scope of this test beyond newborn screening for FXS.
Materials and Methods
Participants and sample processing
The sex chromosome aneuploidy cohort included 257 participants aged between 19 and 69 years. Saliva and venous blood were collected from 162 and 95 individuals, respectively. The fragile X cohort included 588 participants aged between 1 day (newborns) and 67 years, with venous blood (n = 490) and newborn and adult dried blood spots (n = 98) collected from multiple centers in Australia and the United States as detailed in Supplementary Tables S1 and S2 online.
The study was approved by the Royal Children’s Hospital Human Research Ethics Committee and the Southern Health and Monash University Human Research Ethics Committees, both located in Victoria, Australia, and by the Institutional Review Board of the University of California, Davis.
The adult dried blood spots were made at the time of venous blood collection by dropping 50 µl of venous blood onto one disk of a Watman 903 card, and were stored at room temperature for 1 year. The de-identified newborn blood spots (NBSs) from controls and FM individuals were obtained as residual blood spot material through the Victorian Clinical Genetics Services newborn screening program.
One or two 3-mm punches from each dried blood spot disk were collected into 96-well plates or 1.5-ml eppendorf tubes using the Wallac DBS Puncher (Perkin Elmer, Waltham, MA) and stored at room temperature, as indicated in the results section. Each punch or set of punches was incubated in 55 µl of salt lysis buffer (Murdoch Childrens Research Institute, Melbourne, Australia) for 15 min at 98 °C in a heat block for cell lysis, degradation of proteins and release of the DNA into the solution. After boiling, the samples were centrifuged and the supernatant transferred to a fresh 96-well plate. This extract was then treated with sodium bisulfite as described for venous blood and saliva DNA.
The individuals with sex chromosome aneuploidy who provided saliva were recruited from across Australia as part of another study.3 Blood (3–10 ml per individual) was collected in EDTA-treated tubes and DNA extracted using NucleoSpin Tissue genomic DNA extraction kit, as per manufacturer’s instructions (Düren, Germany).
Molecular studies
DNA samples and dried blood spot extract were tested in this study for FMR1 CGG expansion size25 and for methylation via the Sequenom EpiTYPER system as previously described.11 Methylation analysis for each sample was performed in duplicate (unless otherwise stated), giving two separate methylation output ratios (MORs), reflecting technical variation resulting from bisulfite conversion, PCR, and mass cleave reactions.
The 7900HT Fast Real Time PCR machine (Applied Biosystems, Foster City, CA) was used to quantify the sex-determining region Y (SRY) gene copy number, using real-time PCR relative standard curve method,26 normalized to β-globin copy number. The SRY and β-globin dynamic linear range was performed on a series of doubling dilutions of a DNA standard (100–0.8 ng/µl). Previously published sequences were used for primers and probes for the SRY and β-globin genes.27 All samples were assayed in duplicate 10-µl multiplexed PCR reactions. The samples were quantified in arbitrary units (au) in relation to the standard curves performed on each plate.
Data analysis
The sensitivity was considered to be a measure of the probability of correctly identifying the presence of a specific abnormal epi-genotype, i.e., FXS/FM hypermethylated by Southern blot or sex chromosome aneuploidy, with methylation above a methylation threshold of 0.435 or 0.38 as indicated. The specificity was regarded as a measure of the probability of correctly identifying an individual who does not have this epi-genotype, i.e., healthy controls, PM, and unmethylated “high-functioning” FM (UFM) males.
Comparisons between the groups in the median methylation output ratio were conducted using the nonparametric Mann–Whitney two-sample test. All analyses were conducted using the publicly available R statistical computing package (R Development Core Team 2007 R: ISBN 3-900051-07-0. URL: http://www.r-project.org/; accessed 19 February 2009).
Results
Determining technical variation of CpG10–12 methylation analysis for detection of FXS alleles in high quality DNA from lymphoblast cell lines and archival dried blood spots
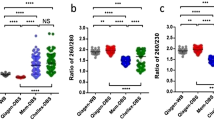
To assess intrarun technical variation on high-quality DNA from patient and control lymphoblast cell lines described in ref. 11, methylation analysis was performed on eight DNA samples on 14 different occasions over a 7-month period by three different operators, with skill sets ranging from experienced to novice. Three of these samples were made by spiking DNA from a healthy male with DNA from an FXS-affected male at different ratios ( Figure 1a ). To assess inter-run variation, we performed analysis on adult dried blood spots (DBSs) from control, PM, and FM males and females, kept at room temperature for 1 year. Of the six individuals, one was a fully affected FM male and another was an asymptomatic female mosaic for PM and FM alleles. The contribution of bisulfite conversion (first stage of analysis) to technical variation in these samples was determined by assaying one DBS from each of the six individuals multiple times with the maximum of nine bisulfite conversions per sample. Each conversion was assessed in triplicate PCR/mass cleave reactions ( Figure 1b ). The contribution of PCR and mass cleave (second stage of analysis) to technical variation in these samples was determined by assaying each of the nine bisulfite conversions either in triplicate or duplicate, or a single PCR/mass cleave reaction ( Figure 1c ). To determine the minimum number of 3-mm blood spot punches required per individuals, we performed CpG10–12 methylation analysis in 40 de-identified control (CGG < 40) NBSs (20 males and 20 females). The comparison indicated that there was no significant difference in MOR between one versus two punches per individual ( Supplementary Table S3 online).
Inter- and intrarun technical variation of FMR1 intron 1 CpG10–12 methylation analysis using MALDI-TOF methylation analysis in adult (>18 years of age) male and female dried blood spots (DBSs) and lymphoblast DNA. (a) To assess intrarun variation, methylation analysis was performed on eight lymphoblast DNA samples on 14 different occasions over a 7-month period by three different operators. Three of these samples were made by spiking DNA from a healthy male with DNA from a fragile X syndrome (FXS)-affected male at 2:1, 1:1, and 1:2 ratios. A curve was generated for the spiked and pure samples to examine the relationship between CpG10–12 methylation output ratios and expected methylation resulting from spiking. (b) To assess inter-run variation resulting from bisulfite conversion (first stage of analysis) and to determine the least number of technical bisulfite conversion replicates that will not make a difference in the result, one DBS from each of the six individuals was assayed multiple times with the maximum of nine bisulfite conversions per sample. Each conversion was assessed in triplicate PCR/mass cleave reactions. Each bar represents the mean of this triplicate assessment for different number of conversions, as indicated. (c) To assess inter-run variation resulting from PCR and mass cleave (second stage of analysis) and to determine the least number of technical PCR and mass cleave replicates that will not make a difference in the result, one DBS from each of the six individuals was bisulfite converted nine times per sample. Each conversion was analyzed either in triplicate, duplicate, or single PCR/mass cleave reaction, as indicated. The error bars represent one SD. FM, full mutation; meth., methylation; MOR, methylation output ratio; PM, premutation.
On the basis of the combined assessment of technical intra and inter-run variation, we conclude that for DBS methylation analysis using this technique, the lowest number of technical replicates required that will make the smallest difference in the output should be one 3-mm punch per individual. This punch should be converted in two bisulfite reactions, with each conversion used for one PCR/mass cleave reaction, and the mean of the two separate reaction representing the final value. It is also important to note that the technical limitation of this approach is that it will not detect a difference in methylation less that 10% of the true value, which is analogous to the contribution of the combined technical variation for FREE1 analysis11; and this limitation should be used when interpreting differences in biological variation.
Comparison between groups: CpG10–12 methylation analysis for detection of FXS alleles in dried blood spots and venous blood
We have previously shown in a pilot cohort that in venous blood the MOR threshold of 0.435 had high specificity and sensitivity (approaching 100%) for detection of FM females with verbal IQ impairment, suggesting that this threshold would be the most appropriate for FXS newborn or early childhood screening.13 In Figure 2 , we show that in a larger cohort, the 0.435 methylation threshold can separate FM individuals from PM and control groups as effectively in 500 ng of DNA from venous blood as in cell extract from one 3-mm punch per sample from dried blood spots of adults and newborns.
Comparison between groups of FMR1 intron 1 CpG10–12 methylation output ratio in 490 venous blood DNA and 98 newborn and adult dried blood spots. (a) Venous blood DNA from 17 control, 38 premutation (PM), and 66 full-mutation (FM) males indentified through the developmental delay/autism spectrum disorder of unknown-cause fragile X syndrome (FXS) testing referrals (formal IQ testing was not performed); as well as four “high-functioning” FM males with verbal IQ > 70 determined to be unmethylated using methylation sensitive Southern blot and identified through cascade testing. (b) De-identified newborn and adult dried blood spots from 20 control, 6 PM, and 20 FM males located through cross-matching laboratory identification numbers of venous blood samples referred for routine fragile X testing; as well as two dried blood spots from “high-functioning” FM males with verbal IQ > 70 determined to be unmethylated using methylation sensitive Southern blot identified through cascade testing. (c) Venous blood DNA from 102 control, 137 PM, and 126 FM females indentified through the developmental delay/autism spectrum disorder of unknown cause and cascade FXS testing (formal IQ testing was not performed). (d) De-identified newborn and adult dried blood spots from 20 control, 6 PM, and 24 FM females located through cross-matching laboratory identification numbers of venous blood samples referred for routine fragile X testing; as well as one Turner syndrome dried blood spot.
In venous blood DNA, all residual de-identified FM male samples obtained from the developmental delay referrals show methylation above the 0.435 threshold, whereas “high-functioning” FM males unmethylated by Southern blot show methylation well below this threshold. Considering that a vast majority of FM males from the developmental delay referrals have FXS, the sensitivity and specificity for FXS in venous blood of this cohort of 125 males are both 100%.
In the NBS and DBS cohort of 48 males, all but one de-identified FM male show methylation above the 0.435 threshold. Unfortunately, there is no clinical information available for the NBS from the unmethylated de-identified FM obtained through cascade testing at Victorian Clinical Genetics Services, as this sample shows MOR equivalent to that of DBSs from the rare “high-functioning” FM males, unmethylated by Southern blot. Therefore, we had to consider this sample as a false negative, resulting in sensitivity of 100% and specificity of 95%, using the 0.435 MOR for methylation analysis in male dried blood spots.
In venous blood DNA from 365 females, 45% of FM individuals showed methylation above the 0.435 MOR, and in dried blood spots from 50 females, the proportion of abnormally methylated FM individuals was 43%, which are consistent with the previously reported 50% frequency of intellectual disability or borderline IQ in FM females.28 We also show that from 346 healthy control and PM males and females, 99% have MORs below 0.435. Therefore, using the 0.435 threshold in this study, we can clearly differentiate hypermethylated FM alleles in males and females from control and PM groups in both venous blood DNA and dried blood spots.
Of the five samples from female control and PM groups that show MORs above 0.435, two are from venous blood DNA of PM females and three are control NBSs ( Figure 2c,d ). All three “false-positive” controls ( Figure 2d in red) were kept at room temperature for >5 years and show MORs >0.435, possibly due to increased DNA fragmentation/poor DNA quality; and thus, if only blood spots that were kept at room temperature for <5 years are included in the analysis, the specificity will be 100%. Of note, a 45,X adult single 3-mm DBS punch showed methylation equivalent to that of male controls ( Figure 2d ).
Comparison between groups: sensitivity and specificity for sex chromosome aneuploidy screening using CpG10–12 methylation and SRY copy number analyses
In this study, none of the saliva DNA samples showed CpG10–12 MOR values above 0.435 ( Figure 3a ). However, for blood DNA, 10 sex chromosome aneuploidy samples showed CpG10–12 MOR values above this threshold ( Figure 4a ), and all of these samples had three or more X chromosomes. When classified into groups, two of two 48,XXXY, three of four 49,XXXXY, and five of eight 47,XXX samples showed values above the MOR of 0.435. Both the 49,XXXXY and 47,XXX groups had methylation that was significantly higher than that of the 46,XX female controls ( Figure 4a ), with only one point of overlap with the control group at its highest MOR value of 0.38, suggesting that having two or more copies of an inactivated X chromosome increases the CpG10–12 MOR above the normal range and the 0.435 MOR threshold.
Comparison between groups of FMRI intron 1 CpG10–12 methylation output ratio (MOR) and sex-determining region Y (SRY ) copy number in 162 saliva DNA samples from control males and females and sex chromosome aneuploidy individuals 19–69 years of age. The sample consisted of 14 46,XY control males (<40 CGGs); 83 46,XX control females (<40 CGGs); 42 47,XXY Klinefelter syndrome males (confirmed by microarray analysis); 13 47,XXY treated as Klinefelter syndrome males (confirmed by androgen receptor methylation analysis and androgen receptor CAG repeat length heterozygosity; no karyotype available); 6 mosaics for 46,XY/47,XXY (confirmed by microarray analysis); and 4 46,XX males with an SRY translocation (confirmed by microarray analysis). (a) CpG10–12 MOR values determined using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. (b) SRY/β-globin copy number ratios determined using real-time PCR.Each blue dot represents the output for a single sample; the box plots represent the median and interquartile range for each group; the red broken line represents the optimal threshold value for detection of full mutation females with verbal IQ < 70; the green broken line represents the optimal threshold value for detection of presence of X-inactivation; and the blue broken line represents the optimal threshold value for detection of presence of one or more copies of Y chromosome.
Comparison between groups of FMR1 intron 1 CpG10–12 methylation output ratio (MOR) and sex-determining region Y ( SRY ) copy number in 95 blood DNA samples from control males and females and sex chromosome aneuploidy individuals 19–69 years of age. The sample consisted of 19 46,XY control males (<40 CGGs); 35 46,XX control females (<40 CGGs); 10 47,XXY Klinefelter syndrome males (karyotype confirmed); 1 48,XXYY male (karyotype confirmed); 4 47,XYY males (karyotype confirmed); 1 48,XXYY/47,XXY mosaic male (karyotype confirmed); 2 48,XXXY males (karyotype confirmed); 4 49,XXXXY males (karyotype confirmed); 8 47,XXX females (karyotype confirmed); and 11 45,X Turner syndrome females (karyotype confirmed). (a) CpG10–12 MOR values determined using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. (b) SRY/β-globin copy number ratios determined using real-time PCR. Each blue dot represents the output for a single sample; the box plots represent the median and interquartile range for each group; the red broken line represents the optimal threshold value for detection of full mutation females with verbal IQ < 70; the green broken line represents the optimal threshold value for detection of presence of X-inactivation; and the blue broken line represents the optimal threshold value for detection of presence of one or more copies of Y chromosome. Comparison with 46,XY controls, ##P < 0.01, ###P < 0.001; comparison with 46,XX controls, **P < 0.01, ***P < 0.001.
For both saliva and blood, the CpG10–12 MOR values were significantly elevated in 46,XX controls and sex chromosome aneuploidy samples as compared with 46,XY controls (P < 0.0001), with no overlap in methylation values above the optimal threshold of 0.11 for detection of X-inactivation/extra copies of the X chromosome. In contrast, the SRY/β-globin copy number ratios were significantly elevated in 46,XY controls and sex chromosome aneuploidy samples, with at least one copy of Y chromosome determined using molecular karyotyping, as compared with 46,XX controls (P < 0.0001) ( Figure 3b , saliva and Figure 4b , blood). There were no SRY assay false negatives showing overlap in values for the 46,XX control range below the optimal threshold value of 0.01 for detection of Y chromosome copy number in both saliva and blood DNA. One of 11 individuals with 45,X Turner syndrome had SRY/β-globin copy number ratio above the 0.01 threshold for the presence of Y chromosome, indicating presence of low level of 45,X/46,XY mosaicism, which was not identified using molecular karyotyping.
When the CpG10–12 MOR and SRY copy number analyses were combined, the true negatives were considered as samples positive for SRY and negative for CpG10–12 MOR (male controls) or negative for SRY and positive for CpG10–12 MOR (female controls). The true positives for sex chromosome aneuploidy were considered as samples either positive for or negative for both SRY and CpG10–12 MOR. Thus, of the 162 saliva DNA samples, 97 were considered to be true negatives and 65 were considered to be true positives. In this context, combined SRY and CpG10–12 MOR analyses for sex chromosome aneuploidy showed sensitivity and specificity of 100% in saliva DNA.
In blood, using the same criteria, 54 of the 95 DNA samples were considered to be true negatives and 37 were considered to be sex chromosome aneuploidy true positives. In blood, only three sex chromosome aneuploidy samples (all with three or more X chromosomes) and one 45,X Turner syndrome sample (low level 45,X/46,XY mosaic) were considered to be false negatives using this approach. Thus, at 0.435 MOR threshold, the combined CpG10–12 MOR and SRY analyses for sex chromosome aneuploidy in blood DNA showed sensitivity of 90% and specificity of 100%. If the CpG10–12 MOR threshold is lowered to the upper value of the control range (0.38), the sensitivity and specificity change to 98 and 99%, respectively.
Discussion
The case for including FXS testing into newborn screening has relied strongly on the failure to diagnose children early enough to provide information to parents on the risk of recurrence in subsequent pregnancies. Most recently, the case has been strengthened by evidence that early intervention with psychological and educational therapies and treatment with psychotropic drugs can improve symptoms of FXS in affected children.29 Although these are not specific for diagnosis of FXS, there is now evidence emerging of newer mGluR5-based therapies specific for FXS, which show potential to be used as part of an FMR1 promoter methylation-based pharmacogenomic approach.30
Another important advance has been the development of a suitable test1,13,31 with high sensitivity and specificity for epimutation detection and prognosis in affected individuals. Of note, this test does not detect the very common unmethylated, gray-zone, and PM alleles that are associated with currently untreatable, late-onset disorders.
The case for newborn screening for sex chromosome aneuploidy is similar. Intervention therapies used at an early age can benefit individuals with these disorders,7,32,33,34 but existing tests such as molecular karyotyping are not suitable for low-cost, high-throughput use. The ability to combine FREE2 methylation and SRY copy number testing removes the technical barrier to screening for sex chromosome aneuploidy and provides the option to perform “FXS only” screening or combined FXS and sex chromosome aneuploidy screening.
Using the FREE2 methylation test for combined FXS and sex chromosome aneuploidy screening
We have, therefore, investigated which sex chromosome aneuploidies would be detected using the FREE2 methylation test if a Y chromosome marker was included and the positive methylation threshold was lowered to just above the upper range of the threshold of male controls. We found that by including the Y chromosome marker with a 0.435 MOR FXS methylation threshold,13 almost all sex chromosome aneuploidies could be differentiated with sensitivity and specificity approaching 100%. The exceptions were one 49,XXXXY and two 47,XXX individuals who could not be differentiated from control females. However, if the methylation threshold was lowered to the MOR of 0.38, which is the upper MOR value of control females, the test would correctly identify these three cases, but at the expense of misclassifying one of 118 XX control females (i.e., a 0.8% false-positive rate).
Using the FREE2 methylation test for only FXS screening
Incidental detection of sex chromosome aneuploidies has also been recently described as a major impediment to the introduction of FXS newborn screening,1,31 as these are fivefold more prevalent than FXS individuals in the general population and may be asymptomatic. In this study, we show that the FREE2 methylation test is specific for hypermethylated FM alleles, and is as robust in one 3-mm punch from an archival dried blood spot as from venous blood DNA. The test also has a very low level of technical variation (<10%). If the sex of the individual tested is not determined (i.e., no SRY marker is used), the test does not differentiate between healthy control males and females, the vast majority of PM and unmethylated FM “high-functioning” males, and individuals with sex chromosome aneuploidies. This approach, thus, overcomes the concerns raised by Coffee.31 The exceptions are some, but not all, individuals with three or more X chromosomes, who are rare. In this scenario, the test is essentially specific for FXS and avoids the ethical concerns related to incidental detection of sex chromosome aneuploidies through FXS newborn screening strategies performed with other FXS methylation and CGG-based tests.31
Proposed follow-up confirmatory testing for screening without use of an SRY marker
Of the 10 aneuploid samples with CpG10–12 MOR above the 0.435 FXS threshold, five were from 48,XXXY and 49,XXXXY individuals; the other five were 47,XXX. In practice, this would mean that all samples with CpG10–12 MOR above 0.435 would need follow-up testing to distinguish between FXS individuals and those with three or more X chromosomes. Therefore, in the context of newborn screening, we recommend all positives with MOR above 0.435 to be re-tested using microarray molecular karyotyping in venous blood or saliva DNA to differentiate rare infants with three or more X chromosomes from FM individuals affected with FXS. Those individuals with a normal number of X chromosomes but with MOR above 0.435 would then need to be re-tested using venous blood or saliva for the presence of expanded methylated FMR1 alleles using the standard fragile X testing protocols involving CGG sizing and methylation analyses.
Projected costs and estimated benefits of FXS newborn screening
Because the NBS screening protocol proposed in this study has no requirement for DNA extraction, the cost of reagents is analogous to that for other FXS CGG-based PCR tests previously proposed for newborn screening.35 Depending on test volume, the approximate cost for inclusion of FXS into newborn screening using FREE2 methylation analysis would be between $700,000 and $1.5 million for every 100,000 babies tested (i.e., lower cost with greater test volume). On the basis of the previously reported FM frequency range (between 1 in 2,000 and 1 in 3,600 in the general population)14 and the fact that approximately half of all FM females will be identified using the FREE2 test, we estimate that between 21 and 38 FXS-affected individuals (and their families) will be identified for every 100,000 newborns screened.
The societal cost of providing health-care services over the lifetime of an individual with FXS is estimated to be between $350,00036 and $2.5 million.37 Newborn screening for FXS has the potential to reduce the incidence of FXS in the population through identification of at-risk families who would be provided reproductive options. Without making any assumptions in estimating the proportion of families that would avail themselves of these options, a circumspect projection would be that there would be the potential for very significant cost savings in the health-care provision for this condition.
A useful comparison may be made with existing programs that have been implemented worldwide to screen newborns for cystic fibrosis (CF), which like FXS currently has no cure.38,39 Following a positive CF screening result, a second-stage DNA test is performed, which in the Victorian Clinical Genetics Services diagnostic laboratory also uses matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. For every 100,000 babies, CF screening identifies 25 affected individuals with the estimated lifetime cost per individual >$350,000. Similar to the estimates for our FXS test, the combined cost of the CF program including follow-up is ~$700,000 for every 100,000 screened.38,39 Newborn screening for CF aims to achieve an early diagnosis and earlier treatment of symptoms. It also benefits the parents of these children by informing them of reproductive options. Given these similarities, the same arguments can be put forward to support newborn screening for FXS.
Although we do not express our preference for or against combined sex chromosome aneuploidy and FXS newborn screening, at the present time, implementation of an “FXS only” test would be more practical, at least from the perspective of health economics. Although the inclusion of an SRY test is not expensive in itself (~$0.8 per sample), it would identify a large number of individuals (1 in 600 in the general population) who would require follow-up with management protocols, which may prove to be too expensive. However, as more information is collected on the benefits of early intervention, this cost–benefit analysis may change.
In summary, early diagnosis of FXS or sex chromosome aneuploidy through newborn screening programs may improve the development and potential of affected individuals through the use of intervention therapy programs. Full consideration of including these disorders in newborn screening is now unimpeded by the lack of tests suitable for this purpose. The FREE2 methylation test provides accurate diagnosis of FXS and also provides prognostic information and if used in combination with a test for SRY can also detect the vast majority of sex chromosome aneuploidies. The comprehensive cost–benefit analysis, including ethical issues, of testing for these disorders within newborn screening programs can now be fully addressed.
Disclosure
D.E.G. is an inventor on a patent related to the technology described in this article. R.J.H. has received grant funding from Roche, Novartis, Seaside Therapeutics, Forest, and Curemark for treatment studies in fragile X syndrome or autism. She has also consulted with Novartis regarding treatment of fragile X syndrome. The other authors declare no conflict of interest.
References
Coffee B, Keith K, Albizua I, et al. Incidence of fragile X syndrome by newborn screening for methylated FMR1 DNA. Am J Hum Genet 2009;85:503–514.
Bojesen A, Juul S, Gravholt CH . Prenatal and postnatal prevalence of Klinefelter syndrome: a national registry study. J Clin Endocrinol Metab 2003;88:622–626.
Herlihy AS, McLachlan RI, Gillam L, Cock ML, Collins V, Halliday JL . The psychosocial impact of Klinefelter syndrome and factors influencing quality of life. Genet Med 2011;13:632–642.
Nielsen J, Wohlert M . Sex chromosome abnormalities found among 34,910 newborn children: results from a 13-year incidence study in Arhus, Denmark. Birth Defects Orig Artic Ser 1990;26:209–223.
Leggett V, Jacobs P, Nation K, Scerif G, Bishop DV . Neurocognitive outcomes of individuals with a sex chromosome trisomy: XXX, XYY, or XXY: a systematic review. Dev Med Child Neurol 2010;52:119–129.
Tartaglia NR, Howell S, Sutherland A, Wilson R, Wilson L . A review of trisomy X (47,XXX). Orphanet J Rare Dis 2010;5:8.
Freriks K, Timmermans J, Beerendonk CC, et al. Standardized multidisciplinary evaluation yields significant previously undiagnosed morbidity in adult women with Turner syndrome. J Clin Endocrinol Metab 2011;96:E1517–E1526.
Bishop DV, Jacobs PA, Lachlan K, et al. Autism, language and communication in children with sex chromosome trisomies. Arch Dis Child 2011;96:954–959.
Ross JL, Samango-Sprouse C, Lahlou N, Kowal K, Elder FF, Zinn A . Early androgen deficiency in infants and young boys with 47,XXY Klinefelter syndrome. Horm Res 2005;64:39–45.
Tost J, Gut, IG . DNA Methylation Analysis by MALDI Mass Spectrometry. Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2012.
Godler DE, Tassone F, Loesch DZ, et al. Methylation of novel markers of fragile X alleles is inversely correlated with FMRP expression and FMR1 activation ratio. Hum Mol Genet 2010;19:1618–1632.
Godler DE Slater HR, Bui QM, et al. FMR1 intron 1 methylation predicts FMRP expression in blood of female carriers of expanded FMR1 alleles. J Mol Diagn 2011;13:528–536.
Godler DE, Slater HR, Bui QM, et al. Fragile X mental retardation 1 (FMR1) intron 1 methylation in blood predicts verbal cognitive impairment in female carriers of expanded FMR1 alleles: evidence from a pilot study. Clin Chem 2012;58:590–598.
Crawford DC, Meadows KL, Newman JL, et al. Prevalence of the fragile X syndrome in African-Americans. Am J Med Genet 2002;110:226–233.
Hagerman RJ . The physical and behavioural phenotype. In: Hagerman RJ, Hagerman P (eds). Fragile X Syndrome: Diagnosis, Treatment and Research. John Hopkins: Baltimore, MD 2002:3–109.
Verkerk AJ, Pieretti M, Sutcliffe JS, et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell 1991;65:905–914.
Pieretti M, Zhang FP, Fu YH, et al. Absence of expression of the FMR-1 gene in fragile X syndrome. Cell 1991;66:817–822.
Weiler IJ, Greenough WT . Synaptic synthesis of the Fragile X protein: possible involvement in synapse maturation and elimination. Am J Med Genet 1999;83:248–252.
Irwin SA, Swain RA, Christmon CA, Chakravarti A, Weiler IJ, Greenough WT . Evidence for altered Fragile-X mental retardation protein expression in response to behavioral stimulation. Neurobiol Learn Mem 2000;74:87–93.
Fu YH, Kuhl DP, Pizzuti A, et al. Variation of the CGG repeat at the fragile X site results in genetic instability: resolution of the Sherman paradox. Cell 1991;67:1047–1058.
Loesch DZ, Khaniani MS, Slater HR, et al. Small CGG repeat expansion alleles of FMR1 gene are associated with parkinsonism. Clin Genet 2009;76:471–476.
Loesch DZ, Godler DE, Evans A, et al. Evidence for the toxicity of bidirectional transcripts and mitochondrial dysfunction in blood associated with small CGG expansions in the FMR1 gene in patients with parkinsonism. Genet Med 2011;13:392–399.
Bretherick KL, Fluker MR, Robinson WP . FMR1 repeat sizes in the gray zone and high end of the normal range are associated with premature ovarian failure. Hum Genet 2005;117:376–382.
Godler DE, Slater HR, Amor D, Loesch DZ . Methylation analysis of fragile X-related epigenetic elements may provide a suitable newborn screening test for fragile X syndrome. Genet Med 2010;12:595.
Khaniani MS, Kalitsis P, Burgess T, Slater HR . An improved Diagnostic PCR Assay for identification of Cryptic Heterozygosity for CGG Triplet Repeat Alleles in the Fragile X Gene (FMR1). Mol Cytogenet 2008;1:5.
Bustin SA . Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J Mol Endocrinol 2002;29:23–39.
Lo YM, Tein MS, Lau TK, et al. Quantitative analysis of fetal DNA in maternal plasma and serum: implications for noninvasive prenatal diagnosis. Am J Hum Genet 1998;62:768–775.
Rousseau F, Heitz D, Biancalana V, et al. Direct diagnosis by DNA analysis of the fragile X syndrome of mental retardation. N Engl J Med 1991;325:1673–1681.
Hagerman RJ, Berry-Kravis E, Kaufmann WE, et al. Advances in the treatment of fragile X syndrome. Pediatrics 2009;123:378–390.
Jacquemont S, Curie A, des Portes V, et al. Epigenetic modification of the FMR1 gene in fragile X syndrome is associated with differential response to the mGluR5 antagonist AFQ056. Sci Transl Med 2011;3:64ra61.
Coffee B . Commentary on population screening for fragile X syndrome. Genet Med 2010;12:411–412.
Pierga JY, Giacchetti S, Vilain E, et al. Dysgerminoma in a pure 45,X Turner syndrome: report of a case and review of the literature. Gynecol Oncol 1994;55(3 Pt 1):459–464.
Samango-Sprouse CA, Gropman AL, Sadeghin T, Kingery M, Lutz-Armstrong M, Rogol AD . Effects of short-course androgen therapy on the neurodevelopmental profile of infants and children with 49,XXXXY syndrome. Acta Paediatr 2010;100:861–865.
Patwardhan AJ, Eliez S, Bender B, Linden MG, Reiss AL . Brain morphology in Klinefelter syndrome: extra X chromosome and testosterone supplementation. Neurology 2000;54:2218–2223.
Tassone F, Pan R, Amiri K, Taylor AK, Hagerman PJ . A rapid polymerase chain reaction-based screening method for identification of all expanded alleles of the fragile X (FMR1) gene in newborn and high-risk populations. J Mol Diagn 2008;10:43–49.
Toledano-Alhadef H, Basel-Vanagaite L, Magal N, et al. Fragile-X carrier screening and the prevalence of premutation and full-mutation carriers in Israel. Am J Hum Genet 2001;69:351–360.
Hollingsworth B, Harris A . Economic evaluation of prenatal population screening for fragile X syndrome. Community Genet 2005;8:68–72.
Castellani C, Picci L, Tamanini A, Girardi P, Rizzotti P, Assael BM . Association between carrier screening and incidence of cystic fibrosis. JAMA 2009;302:2573–2579.
Wells J, Rosenberg M, Hoffman G, Anstead M, Farrell PM . A decision-tree approach to cost comparison of newborn screening strategies for cystic fibrosis. Pediatrics 2012;129:e339–e347.
Acknowledgements
We thank the study participants and their families for their contribution. We also thank Alison Archibald for recruitment and collection of saliva DNA samples from male and female controls, and James Pitt, Nick Tzanakos, and other members of the Newborn Screening Laboratory at the Victorian Clinical Genetics Services for retrieval of residual newborn blood spots. We thank Benjamin Ong from the Sequenom Platform Facility at the Murdoch Chlidrens Research Institute for his assistance with MALDI-TOF MS application. We also thank Christopher Boyer of Bio-Link Australia Pty. for critical review of the manuscript. This work was supported by the Victorian government’s Operational Infrastructure Support Program, an NHMRC development grant (no. 1017263 to H.R.S. and D.E.G.), the E.W. Al Thrasher Award, USA (to H.R.S. and D.E.G.), a National Institute of Child Health and Human Development grant, USA (HD36071 to D.Z.L. and R.J.H.), Andrology Australia, and Monash University and in part by a grant from the South Carolina Department of Disabilities and Special Needs.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Table S1.
(DOC 49 kb)
Supplementary Table S2.
(DOC 48 kb)
Supplementary Table S3.
(DOC 78 kb)
Rights and permissions
About this article
Cite this article
Inaba, Y., Herlihy, A., Schwartz, C. et al. Fragile X–related element 2 methylation analysis may provide a suitable option for inclusion of fragile X syndrome and/or sex chromosome aneuploidy into newborn screening: a technical validation study. Genet Med 15, 290–298 (2013). https://doi.org/10.1038/gim.2012.134
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gim.2012.134
Keywords
This article is cited by
-
DNA methylation and behavioral dysfunction in males with 47,XXY and 49,XXXXY: a pilot study
Clinical Epigenetics (2021)
-
Abnormally Methylated FMR1 in Absence of a Detectable Full Mutation in a U.S.A Patient Cohort Referred for Fragile X Testing
Scientific Reports (2019)
-
Intragenic DNA methylation in buccal epithelial cells and intellectual functioning in a paediatric cohort of males with fragile X
Scientific Reports (2018)
-
Brain structure and intragenic DNA methylation are correlated, and predict executive dysfunction in fragile X premutation females
Translational Psychiatry (2016)