Abstract
Purpose: The aim of the study was to determine the actual GJB2 and GJB6 mutation frequencies in North America after several years of generalized testing for autosomal recessive nonsyndromic sensorineural hearing loss to help guide diagnostic testing algorithms, especially in light of molecular diagnostic follow-up to universal newborn hearing screening.
Methods: Mutation types, frequencies, ethnic distributions, and genotype-phenotype correlations for GJB2 and GJB6 were assessed in a very large North American cohort.
Results: GJB2 variants were identified in 1796 (24.3%) of the 7401 individuals examined, with 399 (5.4%) homozygous and 429 (5.8%) compound heterozygous. GJB6 deletion testing was performed in 12.0% (888/7401) of all cases. The >300-kb deletion was identified in only nine individuals (1.0%), all of whom were compound heterozygous for mutations in GJB2 and GJB6. Among a total of 139 GJB2 variants identified, 53 (38.1%) were previously unreported, presumably representing novel pathogenic or benign variants.
Conclusions: The frequency and distribution of sequence changes in GJB2 and GJB6 in North America differ from those previously reported, suggesting a considerable role for loci other than GJB2 and GJB6 in the etiology of autosomal recessive nonsyndromic sensorineural hearing loss, with minimal prevalence of the GJB6 deletion.
Similar content being viewed by others
Main
Hearing loss is one of the most common inherited disorders present at birth and is the most common congenital sensory impairment. In developed countries, approximately 1 in 1000 children is born deaf, and 1 in 300 has hearing loss significant enough to affect development.1 Moreover, an additional 1 in 1000 children develops severe to profound hearing loss in the first two decades of life.2,3 At least 50% of prelingual hearing loss is thought to be genetic in etiology, and approximately 70% of this genetic hearing loss is nonsyndromic, whereas the remaining 30% is associated with one of the >400 clinical syndromes (e.g., Pendred and Usher) in which hearing loss is a component. Finally, although nonsyndromic sensorineural hearing loss (NSNHL) is associated with a variety of inheritance patterns (e.g., autosomal dominant [AD] in 15–20%, X-linked in 2–3%, and mitochondrial in 1%), approximately 80% of NSNHL exhibits autosomal recessive (AR) inheritance. To date, 67 different loci and 23 different genes have been reported to be causally associated with AR-NSNHL (http://webhost.ua.ac.be/hhh/). Despite this significant heterogeneity, up to 50% of AR-NSNHL is associated with mutations in the locus DFNB1 (MIM 220290) on chromosome 13q12, which contains the GJB2 gene (MIM 121011) encoding connexin 26 (Cx26) and the GJB6 gene (MIM 604418), which encodes connexin 30 (Cx30).4 Cx26 and Cx30 belong to a family of transmembrane proteins, which, as homo- and heteromeric hexamers, form connexons. Docking between two connexons on adjacent cells forms intercellular gap junctions,5 which are permeable to ions and small metabolites ≤1.2 kDa.6 In the cochlea, Cx26- and Cx30-containing gap junctions are proposed to maintain K+ homeostasis,7 thereby contributing to the efficient generation of action potentials in response to sound. Among individuals with DFNB1-associated AR-NSNHL, 98% is estimated to carry two identifiable mutations in GJB2, whereas 2% are reported to have mutations in both GJB2 and GJB6 (Genetests DFNB1, http://www.genetests.org).
The spectrum of sequence variants in GJB2 varies significantly with ethnicity. For example, 35delG is common among white individuals (carrier rate of 2–4%),8–10 235delC in the Japanese (carrier rate of 1–2%),11,12 167delT in the Ashkenazi Jewish population (carrier rate of 4.0%),13 and V37I in Taiwanese (carrier rate of 11.6%).14 Nonetheless, the cumulative frequency of all GJB2 mutations is sufficiently high in most populations to warrant clinical testing, as evidenced by its inclusion in clinical practice guidelines.15 Perhaps more controversial, however, is the role of clinical testing for GJB6 mutations in GJB2 heterozygotes. Although preliminary studies reported that the del(GJB6-D13S1830) mutation explained hearing loss in as many as 30% to 70% of affected GJB2 heterozygotes,16 subsequent studies in other populations clearly suggest that other mutations, both within DFNB1 and elsewhere, contribute significantly to AR-NSNHL.17 To examine actual mutation frequencies in North America after several years of generalized diagnostic and research testing and to help guide diagnostic testing algorithms, especially in light of molecular diagnostic follow-up to universal newborn screening, we conducted a large collaborative study with participation from laboratories in the United States and Canada. This study includes only probands with hearing loss tested in the consortium laboratories and provides a realistic overview of clinical practice and molecular diagnostic testing results in North America.
MATERIALS AND METHODS
Study description
Questionnaires were distributed to 16 facilities participating in the North American Connexin Study Consortium in the United States and Canada. Responding facilities included Children's Hospital of Eastern Ontario, University of Alberta, Case Western Reserve University, Emory University, University of Chicago, University of North Carolina Hospitals, University of Utah School of Medicine, Sacred Heart Medical Center, Columbus Children's Hospital, Cincinnati Children's Hospital, Stanford University Medical Center, Harvard-Partners Center for Genetics & Genomics, Children's Hospital Boston, University of Colorado School of Medicine, Chapman Institute (St. Francis), and Athena Diagnostics. Results from DNA analysis (by polymerase chain reaction [PCR] and/or sequencing) of GJB2 and GJB6 were collected from these facilities. In total, 7401 individuals with hearing loss were evaluated for GJB2 mutations. Among these, 888 (12.0%) were also evaluated for GJB6 mutations (Fig. 1). Although all 16 laboratories performed comprehensive sequencing of GJB2 exon 2, four also sequenced the noncoding exon 1 in a total of 2256 individuals. In addition, 11 of 16 laboratories performed GJB6 testing, in each case testing for the large deletion (GJB6-D13S1830) as previously described,18,19 with one laboratory also sequencing the single coding exon in 112 individuals. Testing for the recently identified 232-kb deletion, del(GJB6-D13S1854),20 was not performed on any patients in this cohort before collection of the study data. Moreover, information about phase was not explicitly provided for any subject in this study. Therefore, we assumed that individuals carrying more than one GJB2 variant were biallelic. In addition to molecular genetic testing results, ethnic and phenotypic information was obtained when available to the participating laboratories. All samples were made anonymous before the data were sent to the Stanford investigators to preclude the ability to retrospectively link a given genotype and/or phenotype to a particular individual. Institutional approval for this study was obtained from Stanford University.
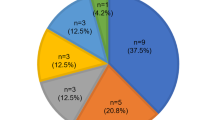
GJB2 and GJB6 testing results. (a) GJB2 results. Het refers to heterozygous individuals with only a single GJB2 variant identified. (b) GJB6 results. Individuals with any GJB6 variant were counted. Nine had the GJB6-D13S1830 deletion, and two had novel variants of unknown relation to hearing loss. All individuals with GJB6 variants were also compound heterozygous for GJB2 variants.
Statistical analysis
Fisher exact tests were performed to determine the statistical significance of differences in prevalence rates between the current and previous studies. Cochran-Mantel-Haenszel tests were performed to evaluate the differences among mutation types (i.e., 35delG versus non-35delG and truncating [T] versus nontruncating [NT]) with respect to the severity of hearing loss (Figs. 2 and 3). Post hoc analysis was performed using the Wilcoxon rank sum test with a Bonferroni correction.
Phenotypes of biallelic GJB2 genotypes, based on presence or absence of 35delG. Individuals were classified according to their reported genotypes (35delG homozygous, 35delG compound heterozygous with a second GJB2 variant, and compound heterozygous with two non-35delG GJB2 variants) and their reported phenotype. Unclassifiable refers to individuals for whom phenotypic information was available but was difficult to categorize. If hearing loss was reported as consecutive ranges (such as mild to moderate), the more severe phenotype was reported. The severity of hearing loss was nonrandomly distributed among the three genotype classes (P = 0.0069, Cochran-Mantel-Haenszel test).
Phenotypes of biallelic GJB2 genotypes. Individuals were classified according to their reported mutation type (two T variants, one T, and one NT variant, or two NT variants) and their reported phenotypes. Unclassifiable refers individuals for whom phenotypic information was available but was difficult to categorize. If hearing loss was reported as consecutive ranges (such as mild to moderate), the more severe phenotype was reported. The degree of hearing loss was nonrandomly distributed among the three genotype classes (P < 0.0001, Cochran-Mantel-Haenszel test).
RESULTS
Demographic and phenotypic information about the study sample is provided in Table 1. In addition to DNA sequencing of the coding exon 2, which was performed by all participating laboratories, four sequenced exon 1 as well. One laboratory, which contributed 112 cases, only reported previously characterized mutations, but not known benign variants. GJB2 variants were identified in 1796 of the 7401 individuals examined (24.3%). Biallelic GJB2 variants were identified in 828 (11.2%) of the 7401 cases, with 399 of these homozygous (48.2%) and 429 compound heterozygous (51.8%) (Fig. 1, a). Among the 828 individuals with biallelic variants, phenotypic information was provided for 215 (26.0%). Seventy-nine of these were homozygous for the 35delG mutation, another 79 (36.7%) were compound heterozygous for 35delG and a second mutation, and 57 (26.5%) had two changes other than 35delG, with severity nonrandomly distributed among these three genotypic classes (Fig. 2; P = 0.0069, Cochran-Mantel-Haenszel test). In 2003, Lim et al.21 reported that the presence of biallelic nonsense mutations correlated with severe to profound hearing loss in the vast majority of cases. Recently, in their study of genotype-phenotype correlations in GJB2-associated hearing loss, Snoeckx et al.22 classified GJB2 variants as either T or NT, concluding that biallelic T mutations were associated with more severe hearing loss. Similar classification in our cohort demonstrated that 59.1% were T/T, 23.7% were T/NT, and 17.2% were NT/NT (vs. 36.7%, 43.8%, and 19.6%, respectively, in the Snoeckx et al. report). Consistent with those studies,21,22 the severity of hearing loss was nonrandomly distributed among these three genotype classes (Fig. 3; P < 0.0001, Cochran-Mantel-Haenszel test), with hearing impairment being more severe in the biallelic T/T class than in the T/NT class, which in turn was more severe than in the NT/NT class.
GJB6 deletion testing was performed in 12.0% (888/7419) of all cases. Of the 888 subjects tested for GJB6, 319 (35.9%) were GJB2 heterozygotes, 115 (13.0%) were GJB2 compound heterozygotes, and 105 (12.0%) were GJB2 homozygotes. No GJB2 variant was identified in 349 (39.3%) of subjects tested for del(GJB6-D13S1830). The >300-kb deletion was identified in only nine individuals (1.0%), all of whom were compound heterozygous for mutations in GJB2 and GJB6 (Fig. 1, b and Table 2). Two novel variants in the GJB6 coding region (689insA and 631T>G) of unclear clinical significance were each reported in one individual. Although the severity of hearing loss among GJB2-GJB6 double heterozygotes ranged from mild to profound, the two cases of del(GJB6-D13S1830) heterozygosity combined with a T GJB2 mutation (frameshift 35delG or W77X) were associated with severe and profound hearing loss, respectively. Unfortunately, the small sample size severely limits the possibility of meaningful genotype-phenotype correlations for GJB6 mutations.
Of the 139 GJB2 variants identified (Table 3), 61 (43.9%) were AR mutations, 5 (3.6%) were AD, 9 (6.5%) represented likely or known benign variants, 11 (7.9%) were reported to be variants with unclear relation to phenotype, and 53 (38.1%) were previously unreported, presumably representing novel mutations or benign variants based on comparison with mutations listed at the Connexin Deafness Homepage (http://davinci.crg.es/deafness) and the Human Gene Mutation Database (www.hgmd.org), in conjunction with PubMed searches (www.pubmed.gov). Overall, the most common mutations across all ethnic groups were 35delG, 101T>C (M34T), 109G>A (V37I), and 167delT, which represented 42.8%, 10.9%, 7.8%, and 3.6% of GJB2 allelic variants, respectively (Table 3). The most common benign variant across all ethnic groups was and 79G>A (V27I, 6.3%). However, as previously reported, the frequency of allelic variants varies significantly with ethnicity (Tables 4 and 5). For example, although 35delG was highly prevalent in white and Hispanic individuals, this variant was not even among the “top 10” in Asians, for whom V37I (109G>A), V27I (79G>A), and E114G (341A>G) predominate. Unfortunately, a relative lack of nonwhite individuals in the study sample preclude stronger conclusions regarding allelic frequencies for these ethnicities. Previously reported dominant mutations were identified in 11 heterozygous individuals (0.1%): four with R75Q (224G>A), three with R184Q (551G>A), two with R75W (223C>T), one with R143Q (428G>A), and one with D50N (148G>A). Concurrent V27I/E114G (79G>A and 341A>G) substitutions, which may have a pathogenic effect when present together in cis,17 were observed in 39 cases (0.5%) with variable degrees of hearing loss. In five of these, a third mutation in GJB2 was reported. (Note that for this study, 79G>A (V27I) and 341A>G (E114G), when present together regardless of their cis/trans relation, were considered a disease-causing mutation, but when present separately, each variant was considered a benign variant.) Of the 828 subjects with biallelic variants, 46 (5.6%) carried at least one variant that was classified as benign, novel, or of unclear clinical significance. Furthermore, the breakdown among the 968 heterozygotes seen in this study according to the classification of the variants shown in Table 3 was 0.9% AD, 74.8% AR, 17.0% benign variant, 3.1% with unknown relationship to disease, and 4.1% novel (data not shown). Finally, unilateral hearing loss was reported in six (0.08%) instances. In three such cases, no GJB2 variant was detected, and GJB6 was not tested. In two instances, the individuals were heterozygous for 35delG and del(GJB6-D13S1830) was not detected. In the final case, the study participant, whose GJB6 status was unknown, was heterozygous for V27I (79G>A).
Among the 53 putatively novel variants (Tables 3, 6, and 7), 40 are nucleotide substitutions (with 36 missense mutations and four synonymous mutations), 7 are frameshift mutations, 4 occur upstream of the transcription initiation site and therefore are of unclear significance, 1 is a nonsense mutation, and 1 is an in-frame deletion. Among the 36 nonsynonymous variants (Tables 6 and 7), 33 occur at residues conserved across the species examined (chimpanzee, mouse, rat, oxen, sheep, and guinea pigs). No obvious ethnic bias was identified among these variants.
DISCUSSION
We present a large study of the frequency and distribution of sequence variations in GJB2 and GJB6 in various ethnic groups in the largest North American study of hearing impaired probands reported to date. The principal weakness of this study was the limited availability of information related to demographics, phenotype, and family history, especially for patients who lacked GJB2 and/or GJB6 variants by current molecular testing methods. More detailed information was uniformly available for patients with GJB2 and/or GJB6 variants. Another obvious limitation is lack of GJB6 results in all patients, but especially in GJB2 heterozygotes, which may result in an inaccurate estimate of the prevalence of the del(GJB6-D13S1830) mutation in our study population. While both are common problems with retrospective, multicenter studies such as ours, much valuable information can still be obtained from such studies.
First and foremost, our findings suggest that the frequency and distribution of sequence changes in GJB2 and GJB6 in North America differ from those previously reported in other studies, although various studies have reported a wide range for the prevalence of DFNB1-associated AR NSNHL,8,9,17,21,23–31 most likely related to ethnic differences between the populations studied. Nonetheless, our results suggest a more prominent role for other loci in the etiology of AR-NSNHL in this patient population. For example, variations in GJB2 were present in individuals with NSNHL in only 11.2% of cases (Fig. 1) in our large North American cohort, even taking into account both AR and AD mutations, as well as those thought to be benign or of unknown clinical significance. This is significantly lower than earlier estimates for the United States, ranging from 21.7%17 to 40%.9 Similarly, the prevalence of GJB6-D13S1830-associated hearing loss (1.0%), which in our study occurred solely in the form of GJB2/GJB6 compound heterozygosity, was far below that previously reported in other North American populations (P = 0.02 compared with Pandya et al.17 and P = 0.0002 compared with del Castillo et al.16). Indeed, a potentially digenic etiology for AR-NSNHL based on concurrent GJB2 and del(GJB6-D13S1830) mutations accounted for hearing loss in only 3.4% of GJB2 heterozygotes tested for del(GJB6-D13S1830) in our study, well below previous estimates ranging from 16% to 21% in two different international studies (P < 0.0001 compared with del Castillo et al.16 and P = 0.0002 compared with Pandya et al.17), including one in North America in which 737 deaf probands were ascertained from the United States and Mexico.17 Although study participants were not tested for the recently described del(GJB6-D13S1854) mutation, the absence of this deletion in 180 GJB2 heterozygotes in Virginia and Iowa suggests that del(GJB6-D13S1854) is unlikely to explain the hearing loss observed in GJB2 heterozygotes, at least in certain populations (such as in the United States, France, and Australia, among others).20 Thus, whereas the frequency of a GJB2 and/or GJB6 etiology may be higher in very well characterized cases of AR-NSNHL, the frequencies reported in this study more accurately reflect the result of clinical practice and diagnostic testing. In addition, the frequency of heterozygosity in this study was 13.1% (Fig. 1). This is markedly above the carrier frequency in a predominantly white population, which has previously been reported to be approximately 3%.9 However, this study consisted solely of probands with NSNHL. Furthermore, the higher frequency of heterozygosity may reflect some previously unclassified autosomal dominant mutations or, perhaps more likely, the presence of as yet unidentified mutations in GJB2 or GJB6. Finally, this finding may be associated with mutations in other genes whose protein products physiologically interact with or influence the function of cochlear gap junctions.
The ethnic distributions of variants were generally similar to published reports.16,17 As shown in Tables 4 and 5, the most frequently observed variants were 35delG in whites and Hispanics, 109G>A (V37I) in Asians, −34C>T in African Americans, and 167delT in Ashkenazi Jews. However, the small number of nonwhites in our study cohort limits our ability to generalize our findings to any other ethnicities. As previously reported in Pandya et al.,17 79G>A (V27I) was observed with a greater frequency in Asians and Hispanics, representing 20.8% and 19.1% of variant alleles seen in these populations, respectively (Table 5). In addition, the 341A>G (E114G) variant was observed most frequently among Asians, representing 17.1% of all variant alleles seen in this population (Table 5 and data not shown). Although the cis/trans relationship was only known for four patients with biallelic V27I/E114G, including one Asian patient with profound hearing loss (ethnicities and phenotypes unknown for the other three), concurrent presence of V27I and E114G (79G>A/341A>G) was observed in 39 instances (0.5% of the 7401 subjects) with a slight Asian predominance and variable degrees of hearing loss (5 profound, 3 severe, 2 moderate, 4 mild, and 3 unclassifiable of the 17 with phenotypic information). In this study, concurrent presence of the V27I and E114G (79G>A/341A>G) variants was considered pathogenic regardless of the confirmed cis/trans status. However, the pathogenicity of V27I/E114G remains controversial, at least in part because V27I/E114G in cis and homozygous V27I/E114G have been reported in unaffected individuals.32 Finally, the 101T>C (M34T) variant, which was observed more frequently than any variant other than 35delG in our study and accounted for 10.9% of all variant alleles across all ethnic groups, was particularly frequent among whites and African Americans. It was homozygous in eight study participants, with variable degrees of hearing loss (three profound, five mild). Taken together with the findings of Snoeckx et al.,22 which demonstrated NSNHL in 38 individuals with the 35delG/M34T genotype and in 16 persons homozygous for M34T, our findings indicate that this variant may not be an innocent variant but rather a pathogenic mutation.
With respect to genotype-phenotype correlations, phenotypic information (by report from the submitting institution) was available for 422 subjects (5.7%), 215 (2.9%) of whom had biallelic GJB2 variants. Analysis of this limited subset demonstrated the following: consistent with recent reports,21,22 genotypic classification of mutations as either T/NT or 35delG/non-35delG were nonrandomly associated with the severity of hearing loss (respectively, P < 0.0001 and P = 0.0069 by Cochran-Mantel-Haenszel test). Moreover, hearing loss was more severe in individuals with biallelic T mutations (T/T) versus those heterozygous for both T and NT mutations (T/NT; P < 0.001 post hoc Wilcoxon rank sum test with Bonferroni correction); in turn, individuals compound heterozygous for T and NT mutations had more severe hearing loss than those with biallelic NT mutations (NT/NT; Fig. 3). However, unlike the study by Snoeckx et al.,22 the difference in the degree of hearing loss between the T/NT and NT/NT classes did not achieve statistical significance in this study (P = 0.71 vs. P < 0.005 in Snoeckx et al. (by post hoc Wilcoxon rank sum test with Bonferroni correction), presumably reflecting the larger sample size in the Snoeckx et al. study.
The classification of mutations (Table 3) is challenging in the absence of functional studies that mimic the in vivo environment. Although nonsense mutations and frameshifts, which often lead to premature termination of translation, are generally accepted as pathogenic due to their obvious effect on the protein, the impact of missense mutations and especially noncoding variants is more difficult to predict. Nevertheless, genotype-phenotype correlations are beginning to emerge.19,22,33 Novel missense mutations can be evaluated for possible pathogenic protein effects by prediction tools such as SIFT (http://blocks.fhcrc.org/sift/SIFT_BLink_submit.html) and PolyPhen (http://coot.embl.de/PolyPhen/). However, these programs are not always in agreement and should be used with caution.32 Our categorization was largely guided by the entries on the Connexin Deafness Homepage and literature searches. However, many variants are rare and have been reported only once or a few times, precluding a general assessment of pathogenicity. For some very recently described sequence variants, specifically 34G>T (G12C) and 40A>G (N14D),32 we adhered to a relatively conservative classification, categorizing these as variants of unclear clinical significance.
Fifty-three putatively novel GJB2 variants with no apparent ethnic biases were identified in the current study (Tables 3, 6, and 7), representing a surprisingly high 38.1% of all variants identified. This illustrates that characterization of variants in this gene is still in progress, although they are often not reported in the literature or in the Connexin Deafness Homepage. It is, therefore, imperative for the development of our understanding of the GJB2 gene, including assessment of pathogenicity and genotype-phenotype correlations, that novel variants be reported to a mutation database and ultimately in the literature, even when they are identified in a clinical molecular diagnostic laboratory rather than in the research setting. Seven of the novel GJB2 variants resulted in frameshifts, 1 produced a premature termination codon, 36 were missense mutations, 4 were synonymous changes, 4 occurred upstream of the transcription initiation site, and 1 was an in-frame deletion of 3 nucleotides. Compared with the nucleotide (data not shown) and protein sequences from other species (Table 6), strict sequence conservation existed at 33 of the 36 residues affected by missense variants. The pathogenic significance of these variants, however, remains to be elucidated by functional studies. To our knowledge, no other mutations or variants have been described at the nonconserved residues (amino acids 105, 159, and 217). The effects of variants upstream of the initiating methionine are unclear, as is the effect of the in-frame deletion. Synonymous changes are unlikely to be pathogenic. Any previously unreported change was classified as novel, including those that involve changes at the nucleotide or amino acid positions of formerly reported variants and those nucleotide changes that result in a previously described amino acid change (Table 3, italics). In such instances, pathogenicity may be similar to that of the change reported earlier, although the replacement of an amino acid by a small or bulky residue may have very different effects on protein function. In the absence of functional studies, missense mutations can be investigated by multiple complementary means: a database and literature search, an evaluation of conservation across species, family studies, and research tools such as SIFT and PolyPhen.
Taken together, our findings suggest that loci other than GJB2 and GJB6 contribute to the pathogenesis of AR-NSNHL and that the full spectrum of GJB2 sequence changes is not yet fully elucidated. In light of our findings, we concur with previous reports that GJB2 testing is warranted in patients with NSNHL. However, the steady decline in the reported prevalence of the del(GJB6-D13S1830) in GJB6 since the initial report from del Castillo et al.16 should give the clinical community in North America pause regarding the utility of screening for this and the recently reported del(GJB6-D13S1854) mutation, even in GJB2 heterozygotes. Nonetheless, given the nominal incremental expense and relative ease of testing for one or both mutations, testing may be appropriate in some GJB2 heterozygotes but should only be considered as a second-tier test. Although nearly all (six of seven with reported ethnic information) GJB6 variants in this study were reported in individuals of white descent, the limited ethnic diversity of the population tested for GJB6 (63.2% were white) prevents us from recommending any stratification of populations for GJB6 testing based solely on ethnicity. The higher frequency of this deletion in Hispanics described in previous publications, however, may guide clinical testing.16 Last, we suggest that consideration be given to testing for SLC26A4, the gene encoding Pendrin and responsible for both NSNHL and syndromic hearing loss (Pendred syndrome), and, depending on clinical and family history, perhaps to a mitochondrial mutation panel in patients with hearing loss for whom no molecular etiology can be defined by testing for the preceding genes.1,34,35 Looking ahead, the advent of universal newborn hearing screening, as well as our growing understanding of the molecular genetics of NSNHL, should facilitate more detailed genotype-phenotype correlations and thereby contribute to improved genetic counseling, prognosis, and management of NSNHL.
References
Schrijver I, Gardner P . Hereditary sensorineural hearing loss: advances in molecular genetics and mutation analysis. Expert Rev Mol Diagn 2006; 6: 375–386.
Mason JA, Herrmann KR . Universal infant hearing screening by automated auditory brainstem response measurement. Pediatrics 1998; 101: 221–228.
Parving A . The need for universal neonatal hearing screening—some aspects of epidemiology and identification. Acta Paediatr Suppl 1999; 88: 69–72.
Kenneson A, Van Naarden Braun K, Boyle C . GJB2 (connexin 26) variants and nonsyndromic sensorineural hearing loss: a HuGE review. Genet Med 2002; 4: 258–274.
Bruzzone R, White TW, Paul DL . Connections with connexins: the molecular basis of direct intercellular signaling. Eur J Biochem 1996; 238: 1–27.
Harris AL, Bevans CG . Exploring hemichannel permeability in vitro. Methods Mol Biol 2001; 154: 357–377.
Kikuchi T, Kimura RS, Paul DL, Adams JC . Gap junctions in the rat cochlea: immunohistochemical and ultrastructural analysis. Anat Embryol (Berl) 1995; 191: 101–118.
Estivill X, Fortina P, Surrey S, Rabionet R, et al. Connexin-26 mutations in sporadic and inherited sensorineural deafness. Lancet 1998; 351: 394–398.
Green GE, Scott DA, McDonald JM, Woodworth GG, et al. Carrier rates in the midwestern United States for GJB2 mutations causing inherited deafness. JAMA 1999; 281: 2211–2216.
Zelante L, Gasparini P, Estivill X, Melchionda S, et al. Connexin26 mutations associated with the most common form of non-syndromic neurosensory autosomal recessive deafness (DFNB1) in Mediterraneans. Hum Mol Genet 1997; 6: 1605–1609.
Abe S, Usami S, Shinkawa H, Kelley PM, et al. Prevalent connexin 26 gene (GJB2) mutations in Japanese. J Med Genet 2000; 37: 41–43.
Kudo T, Ikeda K, Kure S, Matsubara Y, et al. Novel mutations in the connexin 26 gene (GJB2) responsible for childhood deafness in the Japanese population. Am J Med Genet 2000; 90: 141–145.
Morell RJ, Kim HJ, Hood LJ, Goforth L, et al. Mutations in the connexin 26 gene (GJB2) among Ashkenazi Jews with nonsyndromic recessive deafness. N Engl J Med 1998; 339: 1500–1505.
Hwa HL, Ko TM, Hsu CJ, Huang CH, et al. Mutation spectrum of the connexin 26 (GJB2) gene in Taiwanese patients with prelingual deafness. Genet Med 2003; 5: 161–165.
Genetics Evaluation Guidelines for the Etiologic Diagnosis of Congenital Hearing Loss. Genetic Evaluation of Congenital Hearing Loss Expert Panel. ACMG statement. Genet Med 2002; 4: 162–171.
del Castillo I, Moreno-Pelayo MA, del Castillo FJ, Brownstein Z, et al. Prevalence and evolutionary origins of the del(GJB6-D13S1830) mutation in the DFNB1 locus in hearing-impaired subjects: a multicenter study. Am J Hum Genet 2003; 73: 1452–1458.
Pandya A, Arnos KS, Xia XJ, Welch KO, et al. Frequency and distribution of GJB2 (connexin 26) and GJB6 (connexin 30) mutations in a large North American repository of deaf probands. Genet Med 2003; 5: 295–303.
del Castillo I, Villamar M, Moreno-Pelayo MA, del Castillo FJ, et al. A deletion involving the connexin 30 gene in nonsyndromic hearing impairment. N Engl J Med 2002; 346: 243–249.
Lerer I, Sagi M, Ben-Neriah Z, Wang T, et al. A deletion mutation in GJB6 cooperating with a GJB2 mutation in trans in non-syndromic deafness: a novel founder mutation in Ashkenazi Jews. Hum Mutat 2001; 18: 460.
del Castillo FJ, Rodriguez-Ballesteros M, Alvarez A, Hutchin T, et al. A novel deletion involving the connexin-30 gene, del(GJB6-d13s1854), found in trans with mutations in the GJB2 gene (connexin-26) in subjects with DFNB1 non-syndromic hearing impairment. J Med Genet 2005; 42: 588–594.
Lim LH, Bradshaw JK, Guo Y, Pilipenko V, et al. Genotypic and phenotypic correlations of DFNB1-related hearing impairment in the Midwestern United States. Arch Otolaryngol Head Neck Surg 2003; 129: 836–840.
Snoeckx RL, Huygen PL, Feldmann D, Marlin S, et al. GJB2 mutations and degree of hearing loss: a multicenter study. Am J Hum Genet 2005; 77: 945–957.
Dalamon V, Beheran A, Diamante F, Pallares N, et al. Prevalence of GJB2 mutations and the del(GJB6-D13S1830) in Argentinean non-syndromic deaf patients. Hear Res 2005; 207: 43–49.
Gazzaz B, Weil D, Rais L, Akhyat O, et al. Autosomal recessive and sporadic deafness in Morocco: high frequency of the 35delG GJB2 mutation and absence of the 342-kb GJB6 variant. Hear Res 2005; 210: 80–84.
Kalay E, Caylan R, Kremer H, de Brouwer AP, et al. GJB2 mutations in Turkish patients with ARNSHL: prevalence and two novel mutations. Hear Res 2005; 203: 88–93.
Liu XZ, Xia XJ, Ke XM, Ouyang XM, et al. The prevalence of connexin 26 (GJB2) mutations in the Chinese population. Hum Genet 2002; 111: 394–397.
Marlin S, Feldmann D, Blons H, Loundon N, et al. GJB2 and GJB6 mutations: genotypic and phenotypic correlations in a large cohort of hearing-impaired patients. Arch Otolaryngol Head Neck Surg 2005; 131: 481–487.
Propst EJ, Stockley TL, Gordon KA, Harrison RV, et al. Ethnicity and mutations in GJB2 (connexin 26) and GJB6 (connexin 30) in a multi-cultural Canadian paediatric Cochlear Implant Program. Int J Pediatr Otorhinolaryngol 2006; 70: 435–444.
Roux AF, Pallares-Ruiz N, Vielle A, Faugere V, et al. Molecular epidemiology of DFNB1 deafness in France. BMC Med Genet 2004; 5: 5.
Santos RL, Aulchenko YS, Huygen PL, van der Donk KP, et al. Hearing impairment in Dutch patients with connexin 26 (GJB2) and connexin 30 (GJB6) mutations. Int J Pediatr Otorhinolaryngol 2005; 69: 165–174.
Santos RL, Wajid M, Pham TL, Hussan J, et al. Low prevalence of Connexin 26 (GJB2) variants in Pakistani families with autosomal recessive non-syndromic hearing impairment. Clin Genet 2005; 67: 61–68.
Tang HY, Fang P, Ward PA, Schmitt E, et al. DNA sequence analysis of GJB2, encoding connexin 26: Observations from a population of hearing impaired cases and variable carrier rates, complex genotypes, and ethnic stratification of alleles among controls. Am J Med Genet A 2006; 140: 2401–2415.
Huculak C, Bruyere H, Nelson TN, Kozak FK, et al. V37I connexin 26 allele in patients with sensorineural hearing loss: evidence of its pathogenicity. Am J Med Genet A 2006; 140: 2394–2400.
Rehm HL . A genetic approach to the child with sensorineural hearing loss. Semin Perinatol 2005; 29: 173–181.
Robin NH, Prucka SK, Woolley AL, Smith RJ . The use of genetic testing in the evaluation of hearing impairment in a child. Curr Opin Pediatr 2005; 17: 709–712.
Acknowledgements
Margaret Kenna and Heidi Rehm are supported by NIDCD grant R01 DC005248. The authors thank all the individuals at the collaborating facilities who participated in either the acquisition or analysis of the data presented herein, as well as Michael Walker and Brit Turnbull for assistance with the statistical analyses. John C. Carey and Rong Mao are supported in part by the CDC grant M01-RR00064 and University of Utah Clinical Research Center.
Author information
Authors and Affiliations
Corresponding author
Additional information
Disclosure: The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Putcha, G., Bejjani, B., Bleoo, S. et al. A multicenter study of the frequency and distribution of GJB2 and GJB6 mutations in a large North American cohort. Genet Med 9, 413–426 (2007). https://doi.org/10.1097/GIM.0b013e3180a03276
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1097/GIM.0b013e3180a03276
Keywords
This article is cited by
-
The congenital hearing phenotype in GJB2 in Queensland, Australia: V37I and mild hearing loss predominates
European Journal of Human Genetics (2024)
-
Confirmation of COL4A6 variants in X-linked nonsyndromic hearing loss and its clinical implications
European Journal of Human Genetics (2022)
-
Genetic etiology of non-syndromic hearing loss in Latin America
Human Genetics (2022)
-
Undescribed GJB2 c.35dupG homozygous prelingual distinguished from c.35delG homozygous/compound heterozygous deafs, dwelling a German ancestry Venezuelan isolate
Egyptian Journal of Medical Human Genetics (2021)
-
Genetic Susceptibility Study of Chinese Sudden Sensorineural Hearing Loss Patients with Vertigo
Current Medical Science (2021)