Abstract
Purpose: To determine the carrier frequency of the 3199del6 cystic fibrosis (CF) mutation in individuals heterozygous for I148T in a large-scale CF testing population.
Methods: DNA samples from 439 consecutive I148T-heterozygous individuals were screened for the 3199del6 mutation using a laboratory-developed test.
Results: Genotyping revealed four samples heterozygous for the 3199del6 mutation (0.9%). The four samples positive for 3199del6 had an IVS 8 genotype of 7T/9T. The 3199del6 mutation was not observed after genotyping of 348 random, anonymous samples.
Conclusion: The 3199del6 mutation occurs in 0.9% of individuals positive for the I148T mutation and < 0.07% of chromosomes that are wild type for the ACMG panel mutations.
Similar content being viewed by others
Main
Cystic fibrosis (CF) is the most common life-limiting autosomal recessive disease among Caucasians, with an incidence of approximately 1 in 3200.1,2 It also affects smaller proportions of all other ethnic groups and populations. The American College of Medical Genetics (ACMG) and American Society of Obstetricians and Gynecologists (ACOG) have recommended a 25-mutation panel for population-based CF carrier screening. These recommendations used an allele frequency threshold of 0.1% in affected patients for inclusion into the panel. Recent studies demonstrated that I148T, one among the 25-mutation panel, occurs > 100 times more frequently in a carrier-screened population and therefore is probably not a disease-associated mutation.3,4 Subsequently, Rohlfs et al.5 detected a 6-bp deletion, 3199del6, in seven of eight affected patients who were positive for I148T and a classic mutation, but did not detect the deletion in 8 healthy patients with I148T and a classic mutation. The haplotype containing I148T and 3199del6 also included the IVS-8 9T allele. Our data has confirmed that I148T compound heterozygotes are asymptomatic in the absence of 3199del6.3
Thus far, there have been no large prospective studies to determine the frequency of 3199del6 in individuals positive for I148T. Information to date indicates that the 3199del6 mutation is not frequent enough to warrant inclusion in a pan-ethnic CF panel.5 This study analyzed 439 consecutive individuals who tested positive for one copy of the I148T variant, and the data provides the most accurate estimate of the occurrence of the 3199del6 mutation in I148T-positive individuals.
MATERIALS AND METHODS
DNA samples
All samples were submitted to our laboratory for CF DNA analysis. Because the test indication is not routinely provided, we are unable to definitively determine how many tests were requested for carrier screening versus diagnoses. However, based on the reproductive ages of the majority of the tested individuals, carrier screening is the probable indication in most cases. Samples containing I148T for which residual DNA remained were anonymized and analyzed for 3199del6. In addition, 348 consecutive, non-CF screened samples were anonymized and analyzed for 3199del6.
Specimens and DNA extraction
Genomic DNA was extracted from EDTA or ACD-anticoagulated whole-blood specimens using the MagAttract DNA Blood M96 Kit (Qiagen, Foster City, CA) according to the recommendations of the manufacturer.
DNA sequencing
Single exon DNA sequencing was performed as described previously.6
ACMG panel CF testing
CF testing was performed using CF Genotyper Version 3.0 Analyte Specific Reagent (Celera Diagnostics) using an ABI 3100 automated DNA sequencer per the manufacturer's recommendations.
Detection of 3199del6 mutation
A laboratory-developed test using the Promega ReadIT genotyping system was used to detect the 3199del6 mutation. Exon 17a of the CF transmembrane regulator (CFTR) gene was amplified from genomic DNA using the following primers: 5′-TTGTCGCAGTTTTACAACCCTA-3′ (with three 5′ phosphorothioate linkages) and 5′-GTTGCTGTGAGGTTTGGAGGA-3′. A 2-μL aliquot of each sample DNA was added to 11.5 μL 2X PCR Master Mix (Promega Corp, Madison WI) containing 0.50 μmol/L each primer. PCR amplification was performed under the following conditions: one cycle of 5 minutes at 95°C; 31 cycles of 0.5 minutes at 94°C, 0.5 minutes at 62°C, and 1 minute at 72°C; and one cycle of 10 minutes at 72°C, followed by a 4°C hold. All PCR amplifications were performed in a MJR PTC-200 thermocycler (MJ Research, Watertown, MA).
Amplicon interrogation
A 6-μL aliquot of diluted PCR product (1:4 in nuclease-free water) was digested with 25 U bacteriophage T7 Gene 6 exonuclease and 0.5 U calf alkaline intestinal phosphatase at room temperature for 30 minutes. After digestion, 10 μL of 0.06 mol/L NaOH was added to each sample and incubated for 3 minutes. Samples were analyzed in three separate reactions using wild-type (5′-GCTCTCAACATAATAAAAGCCACTAT-3′), mutant (5′-TCTCAACATAATAAAAGCCACTGG-3′), or no probe, respectively. The probes were diluted in 50 mmol/L Tris, 10 mM Mg2Cl to a final concentration of 10 pmol/μL. Interrogation reactions consisted of 10 μL amplicon, 20 μL READIT master mix (2X READIT reaction buffer, 0.2 μmol/L ADP, 2 mmol/L sodium pyrophosphate, 2 U READase Polymerase, 0.1 U READase Kinase), and 100 pmol probe (diluted in 50 mmol/L Tris, 10 mmol/L MgCl). ENLITEN Luciferase Reagent (30 μL, Promega Corp) was then injected into each well. Luminometer data were processed using the READIT Calculator program. Relative response ratios (RRR) were calculated from the raw data to determine genotypes.
RESULTS
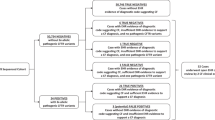
Whenever a sample submitted for analysis was determined to be homozygous or heterozygous for I148T, the referring physician was contacted and urged to request single-exon DNA sequencing for exon 17a to determine if the individual also carried 3199del6. Simultaneously, the automated ReadIT assay was developed, but without a positive genomic control available, the automated assay could not be validated. Eventually, an individual was discovered through single-exon sequencing who carried both 3199del6 and I148T and this sample was then used to validate the automated assay.
Once the assay was optimized, screening for the 3199del6 mutation was performed on all I148T-positive samples for which residual DNA was available. DNA from 439 individuals who were positive for one copy of I148T underwent analysis for the 3199del6 mutation. Of the samples tested, four heterozygotes for the 3199del6 mutation were detected for a rate of 0.9% of this subset of I148T-positive samples. All 3199del6-positive samples had an IVS8 status of 7T/9T, consistent with the observation that 3199del6 occurs solely on I148T/9T chromosomes, although phase could not be established. Because these samples were anonymized, no clinical or ethnic data were available. Analysis of 348 random, anonymized samples submitted for testing other than CF screening did not detect any 3199del6 alleles, indicating that the mutation is found predominantly in the haplotype with I148T/9T.
Before December 2002, we did not store data regarding the IVS8 9T genotype in our database. Since that time, 830 specimens were heterozygous positive for I148T and only 122 (14.7%) were negative for the 9T allele. Although phase cannot be assessed without family studies, it is likely that I148T, like delta F508 occurs primarily in a haplotype including 9T.
DISCUSSION
In a series of 335,204 consecutive CF DNA tests submitted primarily for carrier screening, 1,136 individuals were detected with at least one copy of I148T (data submitted elsewhere). Because 3199del6 occurs with a frequency of 0.9% in individuals heterozygous for I148T, we can estimate that 10 individuals that were positive for I148T would carry 3199del6. Thus, based on a previously-reported CF mutation frequency of 7.7% for the I148T variant,3 3199del6 would account for about 0.07% of CF mutations and would not reach the threshold of 0.1% established by the ACMG. These data suggest that I148T can be removed from the ACMG screening panel.
References
American College of Obstetrics and Gynecology and American College of Medical Genetics. Preconception and prenatal carrier screening for Cystic Fibrosis, clinical and laboratory guidelines. American College of Obstetrics and Gynecology publication: Washington, DC; 2001.
Grody WW, Cutting GR, Klinger KW, Richards CS, Watson M, Desnick RJ . Laboratory standards and guidelines for population based cystic fibrosis carrier screening. Genet Med 2001; 3: 149–154.
Strom CM, Huang D, Buller A, Redman J, Crossley B, Anderson B et al. Cystic fibrosis screening using the college panel: Platform comparison and lessons learned from the first 20,000 samples. Genet Med 2002; 4: 289–296.
Watson MS, Desnick RJ, Grody W, Mennuti MT, Popovich BW, Richards S . Cystic Fibrosis carrier screening: Issues in implementation. Genet Med 2002; 4: 407–409.
Rohlfs EM, Zhou Z, Sugarman EA, Heim EI, Pace RG, Knowles MR et al. The I148T allele occurs on multiple haplotypes: a complex allele is associated with cystic fibrosis. Genet Med 2002; 4: 319–323.
Strom C, Huang S, Chen C, Buller A, Peng M, Quan F et al. Extensive sequencing of the cystic fibrosis transmembrane regulator gene: Assay validation and unexpected benefits of developing a comprehensive test. Genet Med 2003; 5: 9–14.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Buller, A., Olson, S., Redman, J. et al. Frequency of the cystic fibrosis 3199del6 mutation in individuals heterozygous for I148T. Genet Med 6, 108–109 (2004). https://doi.org/10.1097/01.GIM.0000117332.18002.FF
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1097/01.GIM.0000117332.18002.FF
Keywords
This article is cited by
-
Molecular Testing for Cystic Fibrosis Carrier Status Practice Guidelines: Recommendations of the National Society of Genetic Counselors
Journal of Genetic Counseling (2014)
-
Best practice guidelines for molecular genetic diagnosis of cystic fibrosis and CFTR-related disorders – updated European recommendations
European Journal of Human Genetics (2009)
-
Cystic Fibrosis Prenatal Screening in Genetic Counseling Practice: Recommendations of the National Society of Genetic Counselors
Journal of Genetic Counseling (2005)