Abstract
Purpose: To characterize novel mutations in the HEXA gene (α-subunit β-hexosaminidase A).
Methods: Subjects included participants in the California Tay-Sachs disease prevention program. DNA samples from 49 subjects (47 enzymatically defined carriers and 2 disease afflicted) who were negative for the four common disease-associated and the two pseudodeficient mutations, were subjected to single-strand conformation polymorphism (SSCP) analysis over 14 exons.
Results: Targeted sequencing of the 39 electrophoretic variants from SSCP analysis revealed eight novel and deleterious mutations and 31 with previously described mutations. Six novel mutations were found in non-Jewish carriers, and two were found in two patients with infantile Tay-Sachs disease.
Conclusion: Identification of these eight novel mutations provides additional insight to the mutational spectrum for the HEXA gene. Furthermore, this knowledge should enhance diagnosis and prognosis for Tay-Sachs disease, carrier identification, and fundamental studies in structure/function relationships between this gene and its enzymatic product.
Similar content being viewed by others
Main
Tay-Sachs disease (TSD) GM2-gangliosidosis type 1 (MIM 272800) is a progressive neurodegenerative disease that is inherited with an autosomal recessive pattern of inheritance. Mutations that abolish the activity of the A isoenzyme of β-hexosaminidase (EC 3.2.1.52), encoded by the α-subunit HEXA gene, are associated with the lethal infantile form of TSD. Nearly 60 mutant alleles of this type have been described to date.1,2 Population-based carrier screening programs, particularly in the Ashkenazi Jewish population, rely on carrier screening and genetic counseling (including prenatal diagnosis) to reduce the incidence of TSD in this population. In the past three decades, this has accounted for a 90% reduction in the incidence of TSD among Jewish populations in North America.3,4 Carrier screening programs have used the testing of serum and leukocyte hexosaminidase A (Hex A) activity levels and/or DNA-based mutation detection methods as means for carrier identification.
In the general Ashkenazi Jewish population, three mutant alleles account for approximately 93% of enzymatically defined carriers.3,4 These include (1) a TATC insertion in exon 11 (1278insTATC), (2) a G→C transition at the splice junction of exon 12 (IVS12 + 1 G→C I), and (3) a missense mutation in exon 7 (805G→A; G269S) associated with adult-onset GM2 gangliosidosis.4 In non-Jewish individuals, an abnormal splicing mutation in IVS 9 (IVS9 + 1G→A) occurs predominantly in TSD heterozygotes of Celtic heritage.5 Finally, two additional “pseudodeficient” mutations (739C→T; R247W and 745C→T; R249W) have been associated with heterozygous carrier levels of Hex A enzymatic activities.6–8 These “mutations,” however, do not significantly reduce the activity of Hex A for the natural GM2 sphingolipid substrate and, accordingly are of no known biologic consequence.
The California Tay-Sachs Disease Prevention program has tested voluntarily more than 250,000 individuals, of which approximately 50,000 are of non-Jewish ancestry. This results primarily from college-based screenings and other outreach community-based testing activities where all individuals over 18 years of age who choose to be screened are tested. In the California program, all individuals that test as carriers by enzymatic methods are then subjected to DNA testing (examining only the six common mutations described above). With such testing, approximately 98% of Jewish carriers are identified with a known mutation, of which 3% to 4% are a pseudodeficient mutation. Among non-Jewish enzymatically determined carriers using this same DNA test battery, 69% have one of the six mutations identified. However, of these, approximately two thirds carry one of the two pseudodeficient mutations. In the present study, DNA samples from individuals defined as carriers by virtue of heterozygous levels of Hex A enzymatic activity, but who did not demonstrate one of the six common mutations, were subjected to single-strand conformation polymorphism (SSCP) analysis for the possible detection of novel mutations in the HEXA gene. From such an analysis of 49 subjects, 39 were found to have SSCP electrophoretic variants and, of these, 8 novel mutations (disease-associated) were identified.
MATERIALS AND METHODS
Blood samples were drawn from participants in the California Tay-Sachs Disease Prevention program and signed informed consent (institutional review board-approved) was obtained for all subjects. Hex A enzymatic profiles were determined on serum or leukocytes samples as previously described.4 DNA samples from 49 subjects (3 Jewish; 46 non-Jewish) that were negative for both the four common disease-associated and the two pseudodeficient mutations were then analyzed by polymerase chain reaction (PCR)–SSCP.9 DNA was amplified from all 14 exons of the HEXA gene in the presence of 1 μCi of α-32P dCTP (Amersham) using primers and conditions previously described.10 Amplified PCR products were then denatured and subjected to SSCP electrophoresis at room temperature with 0.5× MDE gels (FMC BioProducts) (constant 12 mA at approximately 10,000 Vhr in 0.6 × TBE buffer), and with 5% polyacrylamide (29:1) /5% glycerol (in 0.5 × TBE buffer at a constant 12 mA for approximately 6,000 Vhr). Samples showing band shifts in SSCP gels when compared with normal samples were then directly sequenced using the AmpliCycle sequencing kit (Perkin Elmer). A “control” panel of 56 DNA samples (112 alleles) derived from enzymatically determined noncarriers was screened for the presence of novel mutant alleles. Searches of the NCBI database for HEXA-related sequences were made using the BLAST program,11 accessed at http://www.ncbi.nlm.nih.gov/BLAST/. Multiple sequence alignments were done using the Clustal W program, accessed at http://www.ebi.ac.uk/clustalw/.
RESULTS
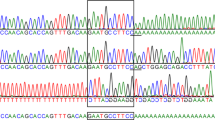
Results of SSCP analysis of DNA samples from 49 subjects revealed 39 electrophoretic band shifts (Table 1). Ten samples (all non-Jewish) showed no SSCP band shifts. Sequencing of exons was carried out on each of the 39 samples with SSCP electrophoretic variations. Thirty-one of the variants (2 Jewish, 29 non-Jewish) were found to have one of 17 previously described mutations. In eight variant samples, novel mutations were identified by DNA sequencing and the biochemical phenotype of these subjects is shown in Table 2. The two subjects with infantile TSD showed virtually no detectable Hex A activity, and the remaining six showed Hex A levels consistent with carrier status. These eight novel mutations are characterized in Table 3 and included two nonsense mutations (Q47X, S52X), two single base pair deletions (1258delT, 1499delT), and four missense mutations (Y37N, H318R, G343 V, and P439S).
These eight novel mutations were not observed in a panel of DNA samples from 56 normal subjects (112 alleles). The two nonsense mutations (Q47X, S52X) were found in two unrelated infants affected with TSD (each a compound heterozygote) (Table 3). Two other mutations (H318R, 1499delT) were identified in two independent obligate carrier parents (Table 3). These four subjects were all non-Jewish and associated with the infantile form of TSD, where a complete deficiency of the Hex A enzymatic activity was found. In the four remaining subjects, one mutation was a stop codon resulting in a frameshift in exon 11, and the other three were missense mutations that are expected to result in nonconservative amino acid changes. Finally, the four missense mutations involved amino acid residues that were evolutionarily conserved across one or more nonhuman species (Table 4).
DISCUSSION
A band shift with SSCP analysis was seen in 79% of subjects in this study (39 of 49 enzymatically defined TSD heterozygotes or disease-afflicted individuals). This rate is consistent with the reported capability of SSCP to detect between 50% and 80% of mutations at a given locus.12 Of the 39 subjects with variant patterns, 31 variants were found to be previously described mutations, and 8 subjects showed novel mutations. Furthermore, these novel mutations were absent in a control population of 56 noncarriers representing 112 alleles. The novel mutations do not appear to disrupt known active sites on the enzyme, sites shown to contain asparagine-linked oligosaccharides, nor are they located at residues that, when mutated, are known to be associated with the B1-variant of TSD.13 The two nonsense mutations in exon 1 (Q47X, S52X) are clearly disease-related mutations, because they were identified in non-Jewish infants of Southwest Hispanic origin whose clinical phenotypes were consistent with classical infantile TSD disease and who were totally deficient in Hex A activity. In a similar manner, the two frameshift mutations (1258delT, 1499delT) are expected to be disease-associated, because virtually all described frameshift mutations in the α subunit HEXA gene are associated with severe infantile TSD.1,2 One of these deletions was seen in Subject 3, the non-Jewish parent of an affected infant, and the other was seen in Subject 6, a carrier by Hex A enzyme activity levels.
The four missense mutations are thought to be disease related, because they were all ascertained in enzymatic heterozygotes or in obligate carriers, and involve nonconservative amino acid substitutions at residues that are conserved across one or more nonhuman species. Previous observations regarding mutations in the α and β subunits genes of the Hex A enzyme suggest that many of the resultant mutant proteins from missense mutations affecting this hexosaminidase are rapidly degraded in the endoplasmic reticulum.13 Furthermore, the amino acid residues at two of the missense mutations are very highly conserved. The novel missense mutations in Subjects 4 and 5 in exons 8 and 9, respectively, (H318R and G343 V) both involve residues that are conserved in bacterial chitobiase (a related glycosyl hydrolase that shows 26% amino acid identity with α-subunit of β-hexosaminidase A14) as well as in Entamoeba histolytica. The H318 residue is also located within four residues of D322, a suspected active site.13 The missense mutation in exon 11 (P439S) in Subject 7 is an amino acid residue conserved in both the mouse and in Entamoeba but not in chitobiase. The Y37 residue in exon 1 in Subject 8 is conserved in the mouse but not conserved in Entamoeba histolytica or in bacterial chitobiase. It is likely that these four missense mutations in the α-subunit of HEXA are associated with abnormal protein retention and degradation within the endoplasmic reticulum, as might be expected.13,15
In summary, identification of these eight novel mutations adds to the extensive mutational spectrum for the α-subunit of the HEXA gene, and contributes to more accurate carrier identification, diagnosis, and prognosis for TSD. In addition, some of these mutations may be relevant to further elucidation of the structure-function relationships of the heterodimeric Hex A (α β) isoenzyme.
References
Myerowitz R . Tay-Sachs disease-causing mutations neutral polymorphisms in the Hex A gene. Hum Mutat 1997; 9: 195–208.
Kaplan F . Tay-Sachs disease carrier screening: a model for prevention of genetic disease. Genet Test 1998; 2: 271–292.
Kaback M, Lim-Steele J, Dabholkar D, Brown D, Levy N . Zeiger K. Tay-Sachs disease-carrier screening, prenatal diagnosis the molecular era. JAMA 1993; 270: 2307–2315.
Kaback M . Population-based genetic screening for reproductive counseling: the Tay-Sachs disease model. Eur J Pediatr 2000; 159( suppl 3): S192–S195.
Landels EC, Green PM, Ellis IH, Fensom AH, Bobrow M . β-Hexosaminidase splice site mutation has a high frequency among non-Jewish Tay-Sachs disease carriers from the British Isles. J Med Genet 1992; 29: 563–567.
Triggs-Raine BL, Mules EH, Kaback MM, Lim-Steele JST, Dowling CE, Akerman BR, Natowicz MR, Grebner EE, Navon R, Welch JP . A pseudodeficiency allele common in non-Jewish Tay-Sachs carriers: implications for carrier screening. Am J Hum Genet 1992; 51: 793–801.
Cao Z, Natowicz MR, Kaback MM, Lim-Steele JST, Prence EM, Brown D, Chabot T, Triggs-Raine BL . A second mutation associated with apparent β-hexosaminidase A pseudodeficiency: identification frequency estimation. Am J Hum Genet 1993; 53: 1198–1205.
Cao Z, Petroulakis E, Salo T, Triggs-Raine B . BenignHEXAmutations, C739T(R247W) C745T (R249W), cause β-hexosaminidase A pseudodeficiency by reducing the α-subunit protein levels. J Biol Chem 1997; 272: 14975–14982.
Orita M, Suzuki Y, Sekiya T, Hayashi K . Rapid sensitive detection of point mutations DNA polymorphisms using the polymerase chain reaction. Genomics 1989; 5: 874–879.
Triggs-Raine BL, Akerman BR, Clarke JTR, Gravel RA . Sequence of DNA flanking exons of theHEXAgene, identification of mutations in Tay-Sachs disease. Am J Hum Genet 1991; 49: 1041–1054.
Altschul SF, Madden TL, Schaffer AA, Zhang J, Jhang Z, Miller W, Lipman DJ . Gapped BLAST PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 1997; 25: 3389–3402.
Cotton RG . Methods in clinical molecular genetics. Eur J Pediatr 2000; 159( suppl 3): S179–S182.
Mahuran DJ . Biochemical consequences of mutations causing the GM2 gangliosidoses [review]. Biochim Biophys Acta 1999; 1455: 105–138.
Tews I, Perrakis A, Oppenheim A, Dauter Z, Wilson KS, Vorgias CE . Bacterial chitobiase structure provides insight into catalytic mechanism the basis of Tay-Sachs disease. Nat Struct Biol 1996; 3: 638–648.
Xie B, Rigat R, Smiljanic-Georgijev N, Deng H, Mahuran D . Biochemical characterization of the Cys138Arg substitution associated with the AB variant form of GM2 gangliosidosis: evidence that Cys138 is required for the recognition of the GM2 activator/GM2 ganglioside complex by β-hexosaminidase A. Biochemistry 1998; 37: 814–821.
Acknowledgements
This research was supported by a contract from the Genetic Disease Branch, Department of Health, State of California.
Author information
Authors and Affiliations
Additional information
Electronic-database information: accession numbers and URL for data in this report are as follows: Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/htbin-post/Omim/dispmim?272800.
Rights and permissions
About this article
Cite this article
Mcginniss, M., Brown, D., Fulwiler, A. et al. Eight novel mutations in the HEXA gene. Genet Med 4, 158–161 (2002). https://doi.org/10.1097/00125817-200205000-00010
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1097/00125817-200205000-00010
Keywords
This article is cited by
-
Biochemical and mutational analyses of HEXA in a cohort of Egyptian patients with infantile Tay-Sachs disease. Expansion of the mutation spectrum
Orphanet Journal of Rare Diseases (2023)
-
Isolation, sequence identification and tissue expression profiles of 3 novel porcine genes:ASPA, NAGA, andHEXA
Journal of Applied Genetics (2008)