Abstract
Purpose: To examine the role of the nuclear genome in affecting the phenotypic expression of the simplest model of a mitochondrial DNA disease, maternally transmitted deafness.
Methods: Linkage analysis in families with maternally inherited deafness associated with the homoplasmic A1555G mutation.
Results: Significant linkage and linkage disequilibrium on chromosome 8 was identified.
Conclusions: This finding represents the first identification of a modifier locus for a human mitochondrial DNA disease and supports the concept of mitochondrial DNA diseases having complex genetic inheritance. The eventual identification of this modifier gene will provide insights into the pathophysiological pathways determining the clinical expression of mitochondrial DNA diseases, an important step toward diagnostic and therapeutic interventions.
Similar content being viewed by others
Main
Mitochondrial DNA diseases are associated with a wide range of human diseases, including systemic and tissue-specific neuromuscular disorders, diabetes mellitus, skin lesions, aplastic anemia, and aging in general.1 Despite the clinical importance of these diseases, and despite the fact that the presumed function of the mitochondrial chromosome have been completely described for nearly two decades, the molecular mechanisms leading from genotype to clinical phenotype remain unknown. This has prevented useful counseling of most patients with mitochondrial DNA mutations, and has also prevented the rational search for therapeutic interventions. To shed light on the pathophysiological pathways between genotype to phenotype, we have concentrated on one of the simplest mitochondrial DNA disorders, the inherited homoplasmic A1555G mutation in the 12S rRNA gene.2 This mutation is associated with nonsyndromic deafness and, despite being homoplasmic, exhibits the same tissue specificity and great variation in clinical expression as the more common heteroplasmic mitochondrial DNA mutations. We have postulated, based on biochemical and genetic analyses, that nuclear-encoded modifier genes are the main explanation for the phenotypic differences within families and that identification of these modifier genes will provide the basis for diagnostic and eventual therapeutic interventions.3–5
Two genome-wide linkage studies in a large Arab-Israeli family and Spanish families with the A1555G mutation led to the conclusion that complex inheritance of nuclear modifier loci is responsible for clinical penetrance of the disease.6,7 Application of nonparametric linkage analysis to the genotyping data of the combined Arab-Israeli, Spanish, and Italian families led to a combined allele-sharing LOD score of 3.1 at locus D8S277, a highly suggestive linkage result.7
To further investigate involvement of this region, we collected an independent set of seven Spanish, Italian, and Finnish families and typed original markers from the chromosome 8 region in all available individuals. Addition of the new families increased the resulting multipoint LOD score from 3.1 to 4.1, thus confirming initial linkage result. Subsequent typing of additional markers and linkage disequilibrium testing revealed significant linkage disequilibrium with D8S277 and two adjacent markers.
SUBJECTS AND METHODS
Pedigrees
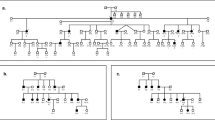
We obtained 47 DNAs from members of five multiplex families from Spain (F1–5), one multiplex family from Italy (F6), and one nuclear family from Finland (F7) with matrilineal nonsyndromic hearing loss (Fig. 1). Each of the families contained at least one pair of affected siblings, and aminoglycoside exposure was noted only in one family (F1) from Spain. Pedigree structures of the Arab-Israeli family, and eight Spanish and two Italian families were described previously.6,7 Informed consent for DNA analysis had been obtained from all members of the Spanish, Italian, and Arab-Israeli families who participated in the study.
Pedigree structure of seven families from Spain (families F1–5), Italy (F6), and Finland (F7). Hearing-impaired individuals are indicated by filled symbols. Asterisks indicate individuals from whom DNA samples were obtained. Filled symbols with opened circles denote individuals who became deaf after aminoglycoside exposure.
Markers
Fluorescently labeled polymerase chain reaction (PCR) primers for markers D8S561 and D8S1469 were purchased from Research Genetics, Inc. (Huntsville, AL). STS sequences corresponding to polymorphic microsatellite repeats D8S518, D8S1798, D8S1742, D8S1819, D8S1140, D8S1825, D8S351 were obtained from the nonredundant GenBank database, and PCR primers were designed using the OLIGO 6.3 program (Molecular Biology Insights, Inc., Cascade, CO). These PCR primers were synthesized with fluorescent tags using the custom oligo synthesis service of Applied Biosystems (Foster City, CA). Polymerase chain reaction was done according to standard protocols in GeneAmp PCR system 9600 (Perkin-Elmer, Norwalk, CT). Electrophoresis of amplified DNA was done on a 377 automated DNA Sequencer (Applied Biosystems). Semiautomated DNA fragment sizing was performed using GENESCAN™ 3.1 software, and genotyping was carried out using GENOTYPER™ 2.5 software (Applied Biosystems).
Linkage analysis
Multipoint nonparametric linkage analysis
To calculate nonparametric multipoint allele-sharing LOD scores, we used the GENEHUNTER-PLUS (ASM 1.0) program.8,9 All the pedigrees included in the analysis were divided into nuclear families, and the scoring function was chosen to be “pairs.” Equal and randomly generated marker allele frequencies were used for the analysis. A genetic map of the region was established combining sex-average map distances reported by the Marshfield Medical Research Foundation with physical map data from the nonredundant (nr) and draft (htgs) GenBank databases.
Transmission Disequilibrium Test
The Transmission Disequilibrium Test10 (TDT) was performed using both the GENEHUNTER 2.0 program5,8 and TDTEX program of the SAGE package release 3.1.11,12 The TDT implemented in the GENEHUNTER 2.0 tests each allele individually and can analyze single marker, as well as two-, three-, and four-marker haplotypes. Permutation test for determining empirical significance levels was implemented. The TDTEX evaluates multialleles simultaneously and can incorporate sibpairs and analyze maternal and paternal transmission separately.13
RESULTS
Linkage analysis of the candidate region around D8S277
We obtained DNA from seven additional families with 47 individuals and 21 affected sib-pairs from Spain, Italy, and Finland (see Fig. 1). First these families were genotyped for the seven markers in the candidate region around marker D8S277, which were used previously to identify the suggestive linkage result in the region.7 These new families were combined with the previously described Arab-Israeli, Spanish, and Italian families for linkage analysis. The resulting maximized allele-sharing LOD score increased from 3.1 in the previously described families to 4.1 (Table 1) in the combined dataset, thus confirming the initial suggestive linkage result.
In order to more precisely localize the putative modifier gene in this region, additional polymorphic markers in the region were typed. A map of the region was established using data from the finished sequence nonredundant GenBank database, as well as from the more preliminary htgs GenBank draft database. We genotyped an additional nine polymorphic microsatellite markers in the candidate region between telomeric marker D8S264 at 1 cM, and centromeric marker D8S503 at 17 cM (D8S277 is at 9 cM) in our existing Arab-Israeli/Spanish/Italian/Finnish family set. The results are shown in Figure 2 and indicate a narrow linkage peak in the region of markers D8S277-D8S561-D8S1819 with a maximum LOD score of 4.0.
According to the recently assembled genomic data in this region, D8S277 and D8S1819 appear to be 300 kb apart. The exact location of the marker D8S561 remains not entirely clear but mostly likely is at 170 kb from D8S277. Same source of genomic data placed next adjacent marker D8S1742, previously considered to be at 3.6 mb away from D8S277, at only 300 kb distance. D8S1742 showed significantly lower multipoint LOD score of 2.1. The adjacent marker D8S1140 on the other side of the peak with a multipoint LOD score of 1.4 still has not been located on the sequence of this region and, therefore, is considered to be 2 cM away according to the linkage maps.
To assess potential variability of the LOD scores depending on marker allele frequencies, we incorporated 10 sets of random marker allele frequencies, as well as CEPH families allele frequencies. The values of the NPL scores and LOD scores were not significantly different from the result using equal marker allele frequencies.
Using the NPL score as the measure of linkage, we observed that nine families had a NPL score < 0, likely unlinked to the chromosome 8 locus, and 19 families had NPL > 0.8, potentially linked to the region. No families with NPL scores between 0 and 0.8 were observed.
Linkage disequilibrium testing of the candidate region by TDT
Each of the markers typed for linkage was also tested for linkage disequilibrium by the TDT using GENEHUNTER 2.0 and TDTEX programs. Using GENEHUNTER 2.0, all three markers with the most significant LOD scores also showed significant linkage disequilibrium with P values as low as 0.0002 for the marker D8S1819 and allele 6 (Table 2). Haplotype analysis did not improve the significance level of the associations. Not all families with NPL scores over 0.8 showed evidence of association with allele 6 of D8S1819.
Exact TDT tests done by TDTEX program analyzing multialleles simultaneously also independently confirmed this statistically significant result for the markers D8S561 (permutation McNemar P value = 0.004) and D8S1819 (P value = 0.005). The results were not significantly different when the analyses were based on affected individual sibs or sibpairs nor when maternal and paternal transmission was analyzed separately or together.
DISCUSSION
Genetic analysis of the previously suggested candidate region around marker D8S277 using additional families provides significant evidence for the first human nuclear modifier locus for maternally inherited deafness. The obtained allele sharing LOD score of 4.1 for the initial markers, and 4.0 for all typed markers, has exceeded the threshold of significant linkage (LOD = 3.6) proposed for genome-wide linkage screens by Lander and Kruglyak.14 Similar results have recently been obtained in mice, when a maternal effect was noted in the (A/J × CAST/Ei) × A/J backcross, with A/J being a hearing-impaired laboratory inbred strain and CAST/Ei being a normal hearing wild-derived inbred strain. The maternal effect was shown to be due to a nucleotide insertion in the mitochondrial tRNA-Arg gene and was only expressed in mice homozygous for the nuclear Ahl locus.15 The Ahl locus had previously been shown to be a major determinant of hearing loss in the A/J strain and a number of other inbred strains.16 The gene responsible for the Ahl locus effect has not yet been identified. Thus, it appears that, both in mouse and man, homoplasmic mitochondrial DNA mutations can cause hearing impairment only in the presence of the appropriate environmental factors17 or nuclear background.
Identification of the gene in the chromosome 8 region will most likely require genetic fine mapping and subsequent candidate gene analysis. For a complex genetic disease, identification of a sharp linkage peak is rare, and the identification of linkage disequilibrium requires often screening of large numbers of markers in large numbers of patients and controls. We were very fortunate that TDT testing revealed highly significant results for several markers in the region and showed that specific preferentially transmitted alleles were identical in the Arab-Israeli and Spanish/Italian families. This is an exciting finding, which may indicate that the modifier gene is in close proximity to these markers. The distance between these markers is 300 kb. The fact that not all chromosome 8 linked families share linkage disequilibrium with the same preferentially transmitted allele is not surprising. First, it should be noted that a NPL score > 0.8 does not necessarily mean that this family is linked to the locus. Second, even if all the families with NPL scores > 0.8 were linked to the chromosome 8 locus, we may not yet have tested the right alleles to see full linkage disequilibrium, or multiple susceptibility mutations may exist. It is also important to emphasize that the chromosome 8 modifier gene is not a disease deterministic gene, but a disease susceptibility gene. That means individuals with this gene will have increased risk for hearing loss, but this gene may not be sufficient to cause the hearing loss. Therefore, it is not surprising to observe some “inconsistent” data. Additional genotyping will be used to define the borders of the linkage disequilibrium region, search for the presence of a susceptibility haplotype, and indicate the most likely region for the modifier gene.
The annotation for the 1 mb contig containing D8S277-D8S561-D8S1819 (accession NT_008268) also contains genes for six known proteins, angiopoietin 2 and five members of the defensin family (defensin alpha 3, alpha 4/corticostatin, alpha 5, alpha 6/Paneth cell-specific, defensin beta 1), as well as one hypothetical protein (FLJ11210). Angiopoietin 2 is an angiogenic factor that is present at the site of vascular remodeling.18 The defensins are antimicrobial peptides that form part of the innate host defense against infection, with alpha-defensins produced in phagocytes and Paneth cells of the small intestine and beta-defensins produced by epithelial cells.19 None of these genes appears to be an obvious candidate for being a modifier gene of the mitochondrial A1555G mutation. The eventual identification of the modifier gene within these regions will shed light on the pathophysiological pathways involved in the clinical expression of the A1555G mutation.
References
MITOMAP. Human Mitochondrial Genome Database. Center for Molecular Medicine Emory University, Atlanta, GA, USA, 2000; http://www.gen.emory.edu/mitomap.html.
Prezant TR, Agapian JV, Bohlman MC, Bu X, Öztas S, Qiu W-Q, Arnos KS, Cortopassi GA, Jaber L, Rotter JI, Shohat M, Fischel-Ghodsian N . Mitochondrial ribosomal RNA mutation associated with both antibiotic-induced and nonsyndromic deafness. Nat Genet 1993; 4: 289–294.
Fischel-Ghodsian N . Mitochondrial mutations and hearing loss: paradigm for mitochondrial genetics. Am J Hum Genet 1998; 62: 15–19.
Guan M-X, Fischel-Ghodsian N, Attardi G . Biochemical evidence for nuclear gene involvement in phenotype of non-syndromic deafness associated with mitochondrial 12S rRNA mutation. Hum Mol Genet 1996; 5: 963–971.
Fischel-Ghodsian N . Homoplasmic mitochondrial DNA diseases as the paradigm to understand the tissue specificity and variable clinical severity of mitochondrial disorders. Mol Genet Metab 2000; 71: 93–99.
Bykhovskaya Y, Shohat M, Ehrenman K, Johnson D, Hamon M, Cantor RM, Aouizerat B, Bu X, Rotter JI, Jaber L, Fischel-Ghodsian N . Evidence for complex nuclear inheritance in a pedigree with nonsyndromic deafness due to a homoplasmic mitochondrial mutation. Am J Med Genet 1998; 77: 421–426.
Bykhovskaya Y, Estivill X, Taylor K, Hang T, Hamon M, Casano RAMS, Yang H, Rotter JI, Shohat M, Fischel-Ghodsian N . Candidate locus for a nuclear modifier gene for maternally inherited deafness. Am J Hum Genet 2000; 66: 1905–1910.
Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES . Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 1996; 58: 1347–1363.
Kong A, Cox NJ . Allele-sharing models: LOD scores and accurate linkage tests. Am J Hum Genet 1997; 61: 1179–1188.
Spielman RS, McGinnis RE, Ewens WJ . Transmission test for linkage disequilibrium: the insulin gene region and insulin-dependent diabetes mellitus (IDDM). Am J Hum Genet 1993; 52: 506–516.
Kruglyak L, Lander ES . Faster multipoint linkage analysis using Fourier transforms. J Comput Biol 1998; 5: 1–7.
S.A.G.E. Statistical analysis for genetic epidemiology, Release 3.1. Computer program available from the Department of Epidemiology and Biostatistics, Rammelkamp Center for Education and Research, MetroHealth Campus. Ed. 3 Cleveland, Case Western Reserve University, 1997.
Cleves MA, Olson JM, Jacobs KB . Exact transmission-disequilibrium tests with multiallelic markers. Genet Epidemiol 1997; 14: 337–347.
Lander ES, Kruglyak L . Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 1995; 11: 241–247.
Johnson KR, Zheng QY, Bykhovskaya Y, Spirina O, Fischel-Ghodsian N . A nuclear-mitochondrial DNA interaction affecting hearing impairment in mice. Nat Genet 2001; 27: 191–194.
Johnson KR, Zheng QY, Erway LC . A major gene on chromosome 10 affecting age-related hearing loss is common to at least ten inbred strains of mice. Genomics 2000; 70: 171–180.
Fischel-Ghodsian N . Genetic factors in aminoglycoside ototoxicity. Ann N Y Acad Sci 1999; 884: 99–109.
Maisonpierre PC, Suri C, Jones PF, Bartunkova S, Wiegand SJ, Radziejewski C, Compton D, McClain J, Aldrich TH, Papadopoulos N, Daly TJ, Davis S, Sato TN, Yancopoulos GD . Angiopoietin-2, a natural antagonist for Tie2 that disrupts in vivo angiogenesis. Science 1997; 277: 55–60.
Van Wetering S, Sterk PJ, Rabe KF, Hiemstra PS . Defensins: key players or bystanders in infection, injury, and repair in the lung?. J Allergy Clin Immunol 1999; 104: 1131–1138.
Acknowledgements
We gratefully acknowledge support by grants RO1DC01402 and RO1DC04092 from the National Institutes of Health/National Institute on Deafness and Other Communication Disorders, Bethesda, MD, and by The Fundacio La Marato de TV3 (XE). Genotyping was supported by NCRR grant MO1 RR00425 to the GCRC phenotyping/genotyping core at Cedars-Sinai Medical Center.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Bykhovskaya, Y., Yang, H., Taylor, K. et al. Modifier locus for mitochondrial DNA disease: Linkage and linkage disequilibrium mapping of a nuclear modifier gene for maternally inherited deafness. Genet Med 3, 177–180 (2001). https://doi.org/10.1097/00125817-200105000-00005
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1097/00125817-200105000-00005
Keywords
This article is cited by
-
The genetic bases for non-syndromic hearing loss among Chinese
Journal of Human Genetics (2009)
-
MRPS18CP2 alleles and DEFA3absence as putative chromosome 8p23.1 modifiers of hearing loss due to mtDNA mutation A1555G in the 12S rRNA gene
BMC Medical Genetics (2007)
-
Lack of a modulative factor in locus 8p23 in a Finnish family with nonsyndromic sensorineural hearing loss associated with the 1555A>G mitochondrial DNA mutation
European Journal of Human Genetics (2003)
-
Two large French pedigrees with non syndromic sensorineural deafness and the mitochondrial DNA T7511C mutation: evidence for a modulatory factor
European Journal of Human Genetics (2002)