Abstract
Purpose: Down syndrome (DS) is a major cause of congenital heart disease (CHD) and the most frequent known cause of atrioventricular septal defects (AVSDs). Molecular studies of rare individuals with CHD and partial duplications of chromosome 21 established a candidate region that included D21S55 through the telomere. We now report human molecular and cardiac data that narrow the DS-CHD region, excluding two candidate regions, and propose DSCAM (Down syndrome cell adhesion molecule) as a candidate gene.
Methods: A panel of 19 individuals with partial trisomy 21 was evaluated using quantitative Southern blot dosage analysis and fluorescence in situ hybridization (FISH) with subsets of 32 BACs spanning the region defined by D21S16 (21q11.2) through the telomere. These BACs span the molecular markers D21S55, ERG, ETS2, MX1/2, collagen XVIII and collagen VI A1/A2. Fourteen individuals are duplicated for the candidate region, of whom eight (57%) have the characteristic spectrum of DS-CHD.
Results: Combining the results from these eight individuals suggests the candidate region for DS-CHD is demarcated by D21S3 (defined by ventricular septal defect), through PFKL (defined by tetralogy of Fallot).
Conclusions: These data suggest that the presence of three copies of gene(s) from the region is sufficient for the production of subsets of DS-CHD. This region does not include genes located near D21S55, previously proposed as a “DS critical region,” or the genes encoding collagens VI and XVIII. Of the potential gene candidates in the narrowed DS-CHD region, DSCAM is notable in that it encodes a cell adhesion molecule, spans more than 840 kb of the candidate region, and is expressed in the heart during cardiac development. Given these properties, we propose DSCAM as a candidate for DS-CHD.
Similar content being viewed by others
Main
Malformations of the cardiovascular system account for the majority of premature deaths caused by congenital anomalies.1 Down syndrome (DS), which is normally caused by trisomy 21,2 is a major cause of congenital heart disease3 and provides an important model with which to link individual genes to the pathways controlling heart development. In this report, we describe an approach to understanding the genes and pathways responsible for congenital heart disease (CHD) in Down syndrome.
The characteristic heart defect seen in DS derives from the abnormal development of the endocardial cushions (EC) and results in a spectrum of defects involving the atrioventricular septum and valves. Accounting for approximately 63% of all DS-CHD,4 these lesions vary in severity from persistence of the common atrioventricular canal and membranous ventricular septum defects to ostium primum patency with valvular anomalies.5,6 The specificity of atrioventricular septal defects (AVSDs) for trisomy 21 is emphasized by the observation that individuals with DS account for 70% of all AVSDs.7 Based both on the aberrant cell adhesion observed for DS cardiac fibroblasts and on subsequent computer modeling that supported the assertion, DS-CHD was proposed to be due to increased expression of molecules involved in cell adhesion.8,9 This hypothesis suggests the existence of a gene or gene cluster on chromosome 21, which is involved in cell adhesion and likely plays an important role in valvuloseptal morphogenesis, but when overexpressed, results in the defects of DS-CHD.
Independent and intersecting approaches to identifying the gene(s) for DS-CHD have included mapping genes known to be involved in cardiac development (none of which localized to chromosome 21), and studying rare individuals with CHD and partial duplications of chromosome 21. These studies initially suggested that subsets of the DS phenotype were associated with three copies of chromosome band 21q22.2–22.3,10–12 and later, that DS-CHD was caused by the overexpression of genes in the region including D21S55 through the telomere.13–15 Subsequent work focused on the identification of a transcriptional map of the DS-CHD region using a 3.5 Mb contiguous clone array covering the interval from D21S55 through MX1/2.16
We now present clinical, cytogenetic, and molecular analyses of a panel of 19 individuals with partial trisomy 21, eight of whom have DS-CHD. Combining the results from these eight individuals suggests a narrowed DS-CHD region including D21S3 through PFKL. The data suggest that three copies of genes in the region of D21S55, previously proposed as a “DS Critical Region,” are not essential for the production of all types of DS-CHD. Moreover, the data do not support an essential role for collagen VI or collagen XVIII in DS-CHD, in that we have identified individuals whose duplications do not include these genes, but nonetheless have CHD of the types found in DS. When combined with evidence showing that deletion of telomeric genes is not associated with CHD,17,18 the data support the hypothesis that trisomy for gene(s) in the DS-CHD candidate region is essential for the production of CHD in DS, although aneuploidy for genes located in the telomeric and other regions likely contributes to the variability of the phenotype.
Only a small number of potential candidates remain in the narrowed candidate region. These include a novel cell adhesion molecule, DSCAM (Down syndrome cell adhesion molecule), which spans more than 840 kb of the candidate region19,20 and is expressed in the heart during cardiac development. Given these properties, we propose DSCAM as a candidate gene for DS-CHD.
METHODS
Clinical analyses
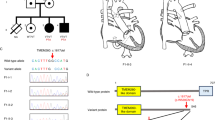
The clinical features of DS in four individuals with partial trisomy 21 were assessed as indicated in Table 1. These data were derived from the family or from original records of the primary care physician, the examining geneticist, or consulting specialists. Full IRB approved procedures were followed and confidentiality was maintained throughout.
Cytogenetic analyses
Extended chromosome preparations were made from peripheral blood lymphocyte cultures by methotrexate synchronization.21 Metaphase-chromosome preparations were made from skin fibroblast cultures using standard procedures. The chromosomes were stained by G-banding (GTG) techniques.22
Molecular analyses
Two methods are used to define the trisomic regions in patients with partial aneuploidy for chromosome 21: quantitative Southern blot dosage analysis and fluorescence in situ hybridization (FISH). Each uses a series of previously mapped chromosome 21 microsatellite DNA markers as probes to define copy number. All probes were isolated as DNA fragments (for Southern blot procedures) or as plasmids, cosmids, or BACs (for FISH studies). The sources and references for all probes used can be found in Table 2.23,24 The probe for ADARB1 was the generous gift of Dr. Katheleen Gardiner at the Eleanor Roosevelt Institute, Denver, CO, and the probes for collagen VI A1 and A2 were the generous gift of Dr. Mon-Li Chu and Dr. Biaggio Saitta at Thomas Jefferson University, Philadelphia, PA.
Genomic DNAs were obtained from fibroblast or lymphoblastoid cell lines. Procedures for DNA isolation and digestion, agarose gel electrophoresis, Southern blot analysis, probe labeling, hybridization, and autoradiogram development were conducted as described previously.13 Southern blots utilized 8–12 paired lanes (16–24 lanes total) of patient and control DNAs. Densitometric analyses utilized the logarithmic transformation of density measurements. FISH studies were conducted using techniques and procedures as described previously.21
RESULTS
Clinical, cytogenetic, and molecular analyses
In order to further narrow the region of chromosome 21 responsible for DS-CHD, we have determined the trisomic regions in a panel of 19 individuals with partial trisomy 21, eight of whom have characteristic DS-CHD. Four of these individuals represent new cases to the literature and exhibit subsets of DS features as described below and in Table 1 (see also Fig. 1). Their chromosome 21 duplications were determined as described in the Methods section, and the results are presented in Table 2 and summarized in Figure 2. The remaining individuals have been reported previously, but their breakpoints have been confirmed or further refined (Figs. 2 and 3, Table 2).
Narrowed DS-CHD candidate region and genes. Solid lines indicate regions of known duplication; open boxes indicate deleted regions. The candidate region is defined as the minimal region of molecular overlap between the individual duplications. Abbreviations denote atrial septal defect (ASD), ventricular septal defect (VSD), atrioventricular septal defect (AVSD), pulmonic stenosis (PS), and tetralogy of Fallot (TOF). The candidate region is denoted by a line with arrows at both ends, with the extent of the region in kilobasepairs indicated at the left of this line. The locations of known genes mapping within the candidate region are indicated by black bars to the left of the gene symbols. Genes mapping within the candidate region include but are not limited to: Down syndrome Critical Region 2 (DSCR2), high mobility group protein 14 (HMG14), tryptophan-rich basic protein (WRB), SH3-binding domain glutamic acid–rich protein (SH3BGR), GlcNAc-beta-1,3-galactosyltransferase 5 (B3GALT5), Purkinje cell protein 4 (PCP4), Down syndrome cell adhesion molecule (DSCAM), beta-site APP-cleaving enzyme 2 (BACE2), myxovirus resistance 1/2 (MX1/2), transmembrane protease serine 2 (TMPRSS2), white protein homolog 1 (ABCG1), trefoil factor 3 (TFF3), trefoil factor 2 (TFF2), trefoil factor 1 (TFF1), human homolog to mouse testis specific gene 2 (TSGA2), cGMP-specific 3′,5′-cyclic phosphodiesterase type 9 (PDE9A), PBX/knotted-1 homeo box-1 (PKNOX1), cystathionine beta-synthase (CBS), alpha crystallin A chain (CRYAA), heat shock transcription factor 2 binding protein (HSF2BP), H2B histone family S member (H2BFS), human pyridoxal kinase (PDXK), cystatin B (CSTB), transmembrane protein 1 (TMEM1), periodic tryptophan protein 2 (PWP2), ES1 protein homolog (HES1), autoimmune regulator (AIRE), and phosphofructo-kinase liver type (PFKL). A complete list of the genes mapping within the region is presented in Hattori et al.20 and at http://www.rzpd.de/general/html/Chrom21/.
Panel of 19 subjects with partial trisomy 21 defines the candidate region for Down syndrome congenital heart disease (DS-CHD). Subjects whose duplications are shown in black have CHD, and those whose duplications are shown in gray do not. The candidate region is defined as the minimal region of molecular overlap among the individuals with partial trisomy 21 and congenital heart disease. Fourteen subjects have duplications that include the DS-CHD region, of whom eight (57%) have CHD. None of the five subjects with duplications outside the region have CHD. Solid lines indicate regions of known duplication; open boxes indicate deleted regions. References for those cases previously reported elsewhere can be found in Table 3 and in Korenberg et al.15 DUP21JL: Park et al.34
Of the eight individuals with DS-CHD, five have aneuploidy for chromosome 21 alone: DUP21BS, DUP21SOL, DUP21SM, DUP21SW, and DUP21ZSC. These individuals exhibit the characteristic spectrum of DS-CHD, which includes atrioventricular septal defects (AVSDs), atrial septal defects (ASDs), ventricular septal defects (VSDs), and tetralogy of Fallot (TOF). Two further cases (DUP21BA and DUP21NA) also carry a small deletion of chromosome 4q35 not associated with CHD, as discussed in Korenberg et al.,12 and the ninth case (DUP21PM) has an inlet type of VSD characteristic of DS and is trisomic for a small region of chromosome 13 in addition to the trisomic region of chromosome 21. Table 3 provides a summary of the karyotypes, chromosome 21 duplications and types of congenital heart disease present in these eight individuals.
Case 1: Patient DUP21BS
This female infant was the product of a full-term gestation born to a 30-year-old G2P2 mother. The pregnancy was complicated by elevated maternal AFP levels (1.26 mg/100 mL), with a risk ratio for DS of 1/227 to 1/149. A paternal grand uncle with DS had congenital heart disease and died at 16 years of age. Amniocentesis carried out at 17 weeks gestation revealed a partial duplication of the long arm of chromosome 21. At birth, DUP21BS exhibited features of DS as listed in Table 1 and illustrated in Figure 1. She did well until 8 hours after birth, when she began to have difficulty with temperature instability and hypoglycemia. She had mild hyperbilirubinemia to 10.9 mg/100 mL and transient thrombocytopenia to 70,000. Cardiac examination at 14 weeks revealed a grade 3/6 slightly harsh systolic ejection murmur and echocardiograms revealed a complete AV canal, consisting of a large ostium primum atrial septal defect (ASD) with a significant left-to-right shunt, and an inlet ventricular septal defect (VSD) effectively closed by a tricuspid pouch. The mitral valve was cleft and mildly regurgitant. There was no patent ductus arteriosus. There was left axis deviation of −60° on EKG, with no evidence of right ventricular hypertrophy. The ASD, VSD, and cleft mitral valve were repaired surgically at 3 years of age, resulting in essentially normal heart function with minimal residual mitral valve insufficiency.
Case 2: Patient DUP21SW
This male infant was the product of a 40-week gestation born to a 27-year-old G1P0 mother and a 24-year-old unrelated father. There is a maternal family history of miscarriages and infant death, but no history of Down syndrome or congenital heart disease.26 DUP21SW was noted at birth to have several facial and visceral features of DS (see Table 1 and Fig. 1). Tetralogy of Fallot was diagnosed by echocardiography within several days of birth. This finding was confirmed at 4 months of age by cardiac catheterization, which revealed normal atrial and systemic pressures in both ventricles, and an aortic O2 saturation of 89%. Cardiovascular examination revealed a grade 3/6 holosystolic murmur best heard at the lower left sternal border. During surgery, his TOF was noted to include an ASD, a large VSD, an overriding aorta with outflow tract stenosis, and a stenotic pulmonic valve. Bars of thickened muscle were excised from the right ventricular outflow tract.
Case 3: Patient DUP21ZSC
This female infant was the product of a full-term gestation born to a 31-year-old G2P2 mother. The pregnancy was uncomplicated. There were multiple variable decelerations during labor, and upon delivery, the infant was given positive pressure ventilation due to persistent cyanosis. She was noted to have several facial features consistent with Down syndrome (Table 1 and Fig. 1). A heart murmur was detected, and an echocardiogram revealed tetralogy of Fallot. This included a large malaligned VSD, displacement of the conal septum, and moderate right outflow tract obstruction due to a stenotic pulmonary valve with an annulus of approximately 4 mm. There was an overriding patent aorta and a small-to-moderate patent ductus arteriosis (PDA) with left-to-right shunting. There was normal systemic and pulmonary venous return to the heart. The aortic valve was tricuspid, with normal right and left coronary arteries and normal branching. The ventricles were well-balanced with good contractility. There was mild hypotonia, no hepatosplenomegaly and normal genitals and extremities. DUP21ZSC was followed to 1 month of age.
Case 4: Patient DUP21PM
This male infant was the product of a 38-week gestation born to a nonconsanguineous couple of Cree Indian descent. The pregnancy was complicated by gestational diabetes type A. His mother had a history of first trimester miscarriages. One sister died of congenital heart disease at 8 months, and another male sibling died at 24 hours. At 5 months of age, congenital heart disease, failure to thrive, and psychomotor retardation were noted. He was marasmic, weighing only 4.35 kg (<3%). In addition to the features of DS noted in Table 1, he had widespaced eyes, frontal bossing, a wide bulbous nose, wide “carp” mouth and high-arched palate, depression of the antihelix bilaterally, mild retrognathism, double left ureter, a small penis, and undescended testes, which were surgically corrected. He also had hyperconvex nails, deep plantar creases and overlapping 2nd over 3rd toes bilaterally. Dermatoglyphics showed normal palmar creases, bilateral distal axial triradii t ′, 10 whorls on fingertips, thenar exit of left A line, loop/loop pattern in left thenar/I1 area, distal loop in left I4, and a small distal loop in the hallucal area bilaterally. Psychomotor development was at the 1-month level. Cardiac catheterizations and surgical repairs at 9 and 15 months demonstrated a large inlet ventricular septal defect located immediately beneath the tricuspid valve with valve tissue overlying much of the defect. This was accompanied by mitral valve insufficiency, a patent ductus arteriosus (PDA), and normal relationship of the great vessels. His psychomotor development remained significantly delayed, and at age 30 months, he could only crawl and make unintelligible sounds. EEG was normal. At 6 years of age, he was incontinent, hyperactive, and had no speech. His gait was wobbly, in part due to bilateral planovalgus feet, and he was barely able to feed himself, although his physical growth was normal—at 6 years his weight was 24.5 kg (75th–90th percentile) and his height was 117 cm (50th percentile).
DISCUSSION
I. Molecular mapping of the DS-CHD candidate region
From clinical, cytogenetic, and molecular analyses, we have determined or refined the chromosome 21 duplications in a panel of 19 individuals with partial trisomy 21, 8 of whom have congenital heart disease (Table 3). We have combined the data from these eight individuals to define a narrowed region of chromosome 21 that is likely to contain the gene(s) responsible for CHD in Down syndrome (Fig. 2).
The validity of a single candidate region for DS-CHD is supported by analysis of all 19 individuals from the partial trisomy 21 panel15 (Fig. 3). Of these, the incidence of CHD in individuals whose duplications include the candidate region (8 of 14 or 57%) is similar to that associated with full trisomy 2113 (50-60%), whereas none of the individuals with duplications outside the candidate region (0 of 5) have CHD (Fig. 3). These data support parsing the DS phenotype into a series of phenotypes, each of which can be affected by different genes.
Narrowing the borders of the DS-CHD region
The telomeric border and the role of collagen VI: The data from DS individual DUP21ZSC, who has TOF, define the telomeric border of the DS-CHD candidate region as PFKL (see Fig. 2). This definition excludes genes located in the subtelomeric region such as S100β,27 as well as ADARB1,28 collagen XVIII,29 and collagen VI A1 and A2, which encode the alpha-1 and alpha-2 chains of collagen VI.30 Collagen VI is a globular heterotrimer expressed in the basement membranes of endothelia and in the endocardial cushions during development,31 which suggested it as a potential candidate for DS-CHD.32 However, DUP21ZSC and two other cases in the literature33 have complex CHD of the types associated with DS and have duplications which do not include collagen VI. Subject LI has TOF associated with duplication from 21pter through CRYA1,33 and subject AL has a VSD associated with duplication extending at least to ITGB2 and excluding collagen VI33 (see Fig. 4). These individuals also carry deletions for the region of collagen VI (Fig. 4), which allows the possibility that decreased expression of collagen VI could be responsible for their heart defects. However, this is not supported by evidence from individuals with isolated deletions of this region. Six such individuals have been reported, none of whom exhibit CHD17,18 (see Fig. 4). There are no cases for comparison in the literature with three copies of the DS-CHD region and two copies of the region of collagen VI. However, we note that DS individual DUP21JL, who is trisomic for the region of collagen VI but has the normal two copies of the DS-CHD candidate region (see Fig. 4), does not have heart disease.34 These data support the hypothesis that three copies of a gene in the proposed DS-CHD candidate region are essential for the production of tetralogy of Fallot or VSD in DS. While aneuploidy for collagen VI and/or other genes in the telomeric region appears to be insufficient to generate DS-CHD, these data provide no evidence for the potential involvement of heterotrisomy for collagen VI genes in DS-CHD. Nonetheless, these genes likely contribute to the phenotypic variability seen in DS-CHD, and we emphasize that the AVSD commonly seen in DS may require three copies of the telomeric region.
DS-CHD is associated with three copies of the candidate region, regardless of copy number for the region of collagen VI. Black lines denote duplicated regions (three copies), gray lines denote normal regions (two copies), and open boxes denote deleted regions (one copy). Four individuals have three copies of the DS-CHD region and one copy of the region of collagen VI, of whom three (75%) have CHD, a percentage similar to that seen in full trisomy 21 (55–65%). Six individuals have two copies of the DS-CHD region and one copy of the region of collagen VI, none of whom have CHD. Individual DUP21JL has three copies of the region of collagen VI and two copies of the DS-CHD region and also does not have CHD. TY, LI, and AL: Pangalos et al.33; MH: Estabrooks et al.18; 4 through 8: McGinness et al.17; JL: Park et al.34 NI, tested but not informative.
The centromeric border: The data from DS individual DUP21PM define the centromeric border of the DS-CHD candidate region as D21S3 (see Fig. 2). This excludes the region of D21S55, previously proposed to be responsible for many of the features of DS10,14,15 and therefore likely excludes genes such as TTC3,35 DYRK1A,36 KCNJ6,37 DSCR4,38 KCNJ15,39 ERG,40 and ETS2.41 DUP21PM carries a 13;21 translocation which also results in trisomy for the short arm and proximal part of the long arm of chromosome 13 (pter-q14) (Table 3). It is, therefore, important to consider whether the heart defect in DUP21PM results from his partial trisomy 21 or his partial trisomy 13. Two lines of evidence support the trisomy of chromosome 21 genes as more likely responsible. First, DUP21PM's heart defect was noted during surgery to be of the type characteristic of DS, a posterior or inlet VSD of the AV canal type, with three small chordae tendineae attached the tricuspid valve cusp around the margin of the defect. Second, although perimembranous VSDs do occur in full trisomy 13, they are associated in 83% (5/6) with a dysplastic tricuspid or pulmonary valve,42 whereas DUP21PM had a normal tricuspid valve which virtually occluded the inlet VSD. There are no cases of pure proximal trisomy 13 with CHD for comparison. Therefore, we infer that the heart defect in DUP21PM is more likely to be due to the overexpression of genes on chromosome 21, although an interaction of genes on 21 and 13 is possible.
It is of interest that, in contrast to his heart disease, many of the relatively nonspecific clinical manifestations in DUP21PM are compatible with the cytogenetic diagnosis of trisomy 13pter-q14, including the microcephaly, wide bulbous nose, wide mouth, retrognathia, undescended testes, and mental retardation.43 Moreover, with the exception of the CHD and mental retardation, the patient had few of the common manifestations of DS.44 The Preus dermatoglyphic diagnostic index for DS45 gave him a score of −1.2 with a 2% probability of having DS. These data support the notion that the features of aneuploidy and DS in particular are modifiable by the combination with other genes but that some features, in this case CHD, may nonetheless be expressed when combined with other aneuploidy.3
The narrowed candidate region for DS-CHD: In summary, combining the results from the eight individuals in our panel with DS-CHD suggests that the candidate region for DS-CHD may be narrowed to D21S3 (defined by VSD), through PFKL (defined by TOF), comprising 5.5 Mb (see Fig. 2). This represents a significant reduction of the previously described candidate region, which spanned 10.5 Mb from D21S55 to the telomere.13,15 The narrowing of the DS-CHD region does not include the region of D21S55, as well as the subtelomeric region and the region of collagen VI (see discussion above). We emphasize that, while these data support the hypothesis that trisomy for a gene in the DS-CHD candidate region is essential for the production of DS-CHD including TOF and VSD, trisomy for additional genes located in the telomeric and other regions likely contributes to the phenotypic variability of DS-CHD.
II. Candidate genes and a hypothesis
DS heart defects characteristically involve the AV septum, suggesting genes expressed in these tissues during development as candidates for DS-CHD. The formation of the AV septum involves the transformation, migration, proliferation, and differentiation of cells from the AV cushions, the primary atrial septum, the dorsal mesocardium, and the neural crest,46 with a possible role indicated for epicardium-derived cells (EPDCs).47 Numerous genes have been identified in these tissues but with the exception of collagen VI A1 and A2, none map to chromosome 21. Therefore, we have used the narrowed DS-CHD candidate region to define potential candidate genes (Fig. 2). The 5.5-Mb region between D21S3 and PFKL contains 39 known genes and 25 predicted genes.20 Potential candidates for DS-CHD were prioritized based first on their expression in the embryonic heart or related tissues, and second on homology to genes known to be involved in AV cushion morphogenesis, particularly in cell adhesion. However, many of the genes in the DS-CHD region are newly identified, and little is known about their expression patterns and/or function. At present, potential candidates known to be expressed in the fetal heart include SH3BGR,48 DSCR249 (described as CHD 1 in Hubert et al.,50 WRB,51 and HES1.52 SH3BGR encodes a glutamic acid–rich protein containing an SH3-binding domain whose expression is ubiquitous but highest in heart and skeletal muscle48 (Lyons and Korenberg, unpublished data); DSCR2 encodes a leucine-rich protein thought to function in cell proliferation, which is expressed in adult heart, skeletal muscle, and other tissues;49 WRB encodes a tryptophan-rich basic protein with a potential nuclear localization signal, which is expressed in adult heart, brain, skeletal muscle, and other tissues, and fetal brain, lung, liver, and kidney;51 and HES1 is thought to function in cellular metabolism and is highly expressed in heart and skeletal muscle.52 Others of the known and predicted genes in the region may be shown to contribute to the DS cardiac phenotype. We now report that DSCAM is also expressed in the fetal heart during development, and discuss evidence which suggests it as a candidate gene for DS-CHD.
DSCAM as a candidate for DS-CHD
The DSCAM gene constitutes a large part of the DS-CHD region, spanning more than 840 kb of the region between D21S3 and PFKL (Fig. 2), as determined from BAC contigs19 and genomic sequence analysis.20 Moreover, it is extraordinarily conserved, with 99% homology between the mouse and human forms at the amino acid level (Korenberg, unpublished data). Northern blot analyses of DSCAM expression showed it to be strongly and specifically expressed in the adult and fetal brain, with lower levels of expression in other tissues.19 Murine fetal tissue in situ hybridization analysis (TISH) on sections of mouse embryos from E8.5 to E17.5 showed abundant DSCAM expression throughout the developing brain, with lower levels of expression in the developing limb bud and in the neural crest and its derivatives, including the cardiac ganglion and the nerves of the mouse fetal heart.19 These results are consistent with the recent finding that the Drosophila DSCAM homolog functions in axon guidance.53 We now report that reverse transcriptase polymerase chain reaction analyses have shown that DSCAM is expressed in the mouse fetal heart at E10.5 p.c., prior to endocardial cushion fusion (Barlow et al., in preparation), and in human fetal heart tissues at 12 weeks of development (Kurnit and Korenberg, in preparation; figures available at http://www.csmc.edu/genetics/korenberg/rt-pcr-dscam.html). Further, both the etiology of characteristic DS heart defects and the increased intercellular adhesiveness of DS cardiac fibroblasts8 implicate disturbances in cell adhesion as underlying DS-CHD. Of the 64 known and predicted genes20 identified in the narrowed DS-CHD candidate region to date, DSCAM is the only one which has been shown to mediate cell-cell adhesion.54 This expression pattern, together with its location within the candidate region and its identity as a cell adhesion molecule of the immunoglobulin (Ig) superfamily (Ig-CAMs), support a potential role for DSCAM in DS-CHD.
To evaluate DSCAM as a candidate for DS-CHD, central questions will include which embryonic heart tissues and developmental processes are primarily affected. Dramatic decreases in the expression of another Ig-CAM, NCAM, occur in cells of the endocardium during epithelial-mesenchymal transformation.55,56 We speculate that the overexpression of DSCAM may have the potential to perturb epithelial-mesenchymal transformation and/or the migration and proliferation of mesenchyme cells, and possibly thus contribute to the increased intercellular adhesion seen in DS cushion fibroblasts8 and the abnormal cushion development seen in DS-CHD. Further questions include whether DSCAM effects are mediated through homophilic adhesion, as suggested by recent studies in mouse fibroblasts54 or through heterophilic adhesion and signaling interactions involving coreceptors and other cushion molecules. Extracellular matrix (ECM) molecules are involved in a large number of interactions, e.g., heparan sulfates bind to a number of different ECM molecules, including NCAM.57 DSCAM also has a region of homology to FGFR4 (Barlow et al., in preparation), which may implicate a role in the regulation of mesenchyme proliferation,46 by analogy to the interactions between Ig domains of NCAM and FGFR1 that contribute to neurite outgrowth.58 The precise mechanisms underlying DSCAM interactions may ultimately be determined through study of DS tissues at the time of differentiation or in a variety of transgenic mouse and other model systems such as the collagen gel explant system55 in which genes can be overexpressed and the effects on morphogenetic processes measured. The results of these investigations should help to elucidate the basis of the most common congenital heart anomalies, defects of the valves and septae, as well as the possible role played by DSCAM and other chromosome 21 genes in generating DS phenotypes of the heart and brain.
References
Clark EB . Mechanisms in the pathogenesis of congenital heart defects. In: Pierpoint MEM, Moller JH, editors. The genetics of cardiovascular disease. Boston: Martinus-Nijoff, 1987: 3–11.
Lejeune J, Gauthier M, Turpin R . Etude des chromosomes somatiques de neuf enfants mongoliens. C R Acad Sci (Paris) 1959; 248: 1721–1722.
Epstein CJ . The consequences of chromosomal imbalance: principles, mechanisms, and models. New York: Cambridge University Press, 1986.
Van Praag R, Papagiannis J, Bar-El YI, Schwint OA . The heart in Down syndrome: pathologic anatomy. In: Marino B, Pueschel SM, editors. Heart disease in persons with Down syndrome. Baltimore, MD: Paul H Brookes Publishing Co., 1996; 69–110.
Cooney TP, Thurlbeck WM . Pulmonary hypoplasia in Down's syndrome. N Engl J Med 1982; 307: 1170–1173.
Anderson RH . Simplifying the understanding of congenital malformations of the heart. Int J Cardiol 1991; 32: 131–142.
Ferencz C, Loffredo CA, Correa-Villasenor A, Wilson PD, editors, Vol 5. Armonk NY: The Baltimore-Washington Infant Study, 1997.
Wright TC, Orkin RW, Destrempes M, Kurnit DM . Increased adhesiveness of Down syndrome fetal fibroblasts in vitro. Proc Natl Acad Sci U S A 1984; 81: 2426–2430.
Kurnit DM, Aldridge JF, Matsuoka R, Matthysse S . Increased adhesiveness of trisomy 21 cells and atrioventricular canal malformations in Down syndrome: a stochastic model. Am J Med Genet 1985; 20: 385–399.
Rahmani Z, Blouin JL, Creau-Goldberg N, Watkins PC, Mattei JF, Poissonnier M, Prieur M, Chettouh Z, Nicole A, Aurias A, Sinet P, Delabar J . Critical role of the D21S55 region on chromosome 21 in the pathogenesis of Down syndrome. Proc Natl Acad Sci U S A 1989; 86: 5958–5962.
McCormick MK, Schinzel A, Petersen MB, Stetten G, Driscoll DJ, Cantu ES, Tranebjaerg L, Mikkelsen M, Watkins PC, Antonarakis SE . Molecular genetic approach to the characterization of the “Down syndrome region” of chromosome 21. Genomics 1989; 5: 325–331.
Korenberg JR, Kawashima H, Pulst SM, Ikeuchi T, Ogasawara SA, Yamatoto K, Schonberg SA . Molecular definition of a region of chromosome 21 that causes features of the Down syndrome phenotype. Am J Hum Genet 1990; 47: 236–246.
Korenberg JR, Bradley C, Disteche C . Down syndrome: molecular mapping of the congenital heart disease and duodenal stenosis. Am J Hum Genet 1992; 50: 294–302.
Delabar JM, Theophile D, Rahmani Z, Chettouh Z, Blouin JL, Preiur M, Noel B, Sinet PM . Molecular mapping of 24 features of Down syndrome on chromosome 21. Eur J Hum Genet 1993; 1: 114–124.
Korenberg JR, Chen X-N, Schipper R, Sun Z, Gonsky R, Gerwehr S, Carpenter N, Daumer C, Dignan P, Disteche C, Graham JM, Hudgins L, McGillivray B, Miyazaki K, Ogasawara N, Park JP, Pagon R, Pueschel S, Sack G, Say B, Schuffenhaur S, Soukup S, Yamanaka T . Down syndrome phenotypes: the consequences of chromosomal imbalance. Proc Natl Acad Sci U S A 1994; 91: 4997–5001.
Hubert RS, Mitchell S, Chen X-N, Ekmekji K, Gadomski C, Sun Z, Noya D, Kim U-J, Chen C, Shizuya H, Simon M, de Jong PJ, Korenberg JR . BAC and PAC contigs covering 3.5 Mb of the Down syndrome congenital heart disease region between D21S55 and MX1 on chromosome 21. Genomics 1997; 41: 218–226.
McGinniss MJ, Kazazian HH, Stetten G, Petersen MB, Boman H, Engel E, Greenberg F, Hertz JM, Johnson A, Laca Z, Mikkelsen M, Patil SR, Schinzel AA, Tranebjaerg L, Antonarakis AE . Mechanisms of ring chromosome formation in 11 cases of human ring chromosome 21. Am J Hum Genet 1992; 50: 15–28.
Estabrooks LL, Rao KW, Donahue RP, Aylsworth AS . Holoprosencephaly in an infant with a minute deletion of chromosome 21(q22.3). Am J Med Genet 1990; 36: 306–309.
Yamakawa K, Huo YK, Haendelt MA, Hubert R, Chen X-N, Lyons GE, Korenberg JR . DSCAM: a novel member of the immunoglobulin superfamily maps in a Down syndrome region and is involved in the development of the nervous system. Hum Mol Genet 1998; 7: 227–237.
Hattori M, Fujiyama A, Taylor TD, Watanabe H, Yada T, Park HS, Toyoda A, Ishii K, Totoki Y, Choi DK, Soeda E, Ohki M, Takagi T, Sakaki T, Taudien S, Blechschmidt K, Polley A, Menzel U, Delabar J, Kumpf K, Lehmann R, Patterson D, Reichwald K, Rump A, Schillhabel M, Schudy A, Zimmermann W, Rosenthal A, Kudoh J, Shibuya K, Kawasaki K, Asakawa S, Shintani A, Sasaki T, Nagamine K, Mitsuyama S, Antonarakis SE, Minoshima S, Shimizu N, Nordsiek G, Hornischer K, Brandt P, Scharfe M, Schön O, Desario A, Reichelt J, Kauer G, Blöcker H, Ramser J, Beck A, Klages S, Hennig S, Riesselmann L, Dagand E, Haaf T, Wehrmeyer S, Borzym K, Gardiner K, Nizetic D, Francis F, Lehrach H, Reinhardt R, Yaspo ML . The DNA sequence of human chromosome 21. Nature 2000; 405: 311–319.
Korenberg JR, Chen X-N . Human cDNA mapping using a high-resolution R-banding technique and fluorescence in situ hybridization. Cytogenet Cell Genet 1995; 69: 196–200.
Mitelman F, editor. ISCN 1995: an international system for human cytogenetic nomenclature. Basel: Krager, 1995.
Korenberg JR, Chen XN, Sun Z, Shi ZY, Ma S, Vataru E, Yimlamai D, Weissenbach JS, Shizuya H, Simon MI, Gerety SS, Nguyen H, Zemsteva IS, Hui L, Silva J, Wu X, Birren BW, Hudson TJ . Human genome anatomy: BACs integrating the genetic and cytogenetic maps for bridging genome and biomedicine. Genome Res 1999; 9: 994–1001.
Antonarakis SE, Patterson D, Van Broeckhoven C, Shimizu N, Gardiner K, Delabar J, Korenberg JR, Reeves R . Report of the committee on the genetic constitution of chromosome 21. In: Cuticchia AJ, Chipperfield MA, Foster PA, editors. Human gene mapping 1995: a compendium. Baltimore and London: The Johns Hopkins University Press, 1996; 1060–1099.
Miyazaki K, Yamanaka T, Ogasawara N . A boy with Down's syndrome having recombinant chromosome 21 but no SOD-1 excess. Clin Genet 1987; 32: 383–387.
Jewett T, Rao PN, Berry M, Pettenati MJ . Down syndrome due to a recombination of a chromosome 21 paracentric inversion in 1 of 2 cases with a review of paracentric recombinants. Am J Hum Genet 1994; 55: A108.
Allore R, O'Hanlon D, Price R, Neilson K, Willard HF, Cox DR, Marks A, Dunn RJ . Gene encoding the beta-subunit of S100 protein is on chromosome 21: implications for Down syndrome. Science 1988; 239: 1311–1313.
Mittaz L, Scott HS, Rossier C, Seeburg PH, Higuchi M, Antonarakis SE . Cloning of a human RNA editing deaminase (ADARB1) of glutamate receptors that maps to chromosome 21q22.3. Genomics 1997; 41: 210–217.
Oh SP, Kamagata Y, Muragaki Y, Timmons S, Ooshima A, Olsen BR . Isolation and sequencing of cDNAs for proteins with multiple domains of Gly-X-Y repeats identify a novel family of collagenous proteins. Proc Natl Acad Sci U S A 1994; 91: 4229–4233.
Weil D, Mattei MG, Passage E, N'Guyen VC, Pribula-Conway D, Mann K, Deutzmann R, Timpl R, Chu ML . Cloning and chromosomal localization of human genes encoding the three chains of type VI collagen. Am J Hum Genet 1988; 42: 435–445.
Klewer SE, Krob SL, Kolker SJ, Kitten GT . Expression of type VI collagen in the developing mouse heart. Dev Dyn 1998; 211: 248–255.
Davies GE, Howard CM, Farrer MJ, Coleman MM, Bennett LB, Cullen LM, Wyse RK, Burn J, Williamson R, Kessling AM . Genetic variation in the COL6A1 region is associated with congenital heart defects in trisomy 21 (Down's syndrome). Ann Hum Genet 1995; 59: 253–269.
Pangalos C, Theophile D, Sinet PM, Marks A, Stamboulieh-Abazis D, Chettouh Z, Prieur M, Verellen C, Rethore M-O, Lejeune J, Delabar J-M . No significant effect of monosomy for distal 21q22.3 on the Down syndrome phenotype in “mirror” duplications of chromosome 21. Am J Hum Genet 1992; 51: 1240–1250.
Park JP, Wurster-Hill DH, Andrews PA, Cooley WC, Graham JM . Free proximal trisomy 21 without the Down syndrome. Clin Genet 1987; 32: 342–348.
Ohira M, Ootsuyama A, Suzuki E, Ichikawa H, Seki N, Nagase T, Nomura N, Ohki M . Identification of a novel human gene containing the tetratricopeptide repeat domain from the Down syndrome region of chromosome 21. DNA Res 1996; 3: 9–16.
Shindoh N, Kudoh J, Maeda H, Yamaki A, Minoshima S, Shimizu Y, Shimizu N . Cloning of a human homolog of the Drosophila minibrain/rat Dyrk gene from ‘the Down syndrome critical region’ of chromosome 21. Biochem Biophys Res Commun 1996; 225: 92–99.
Sakura H, Bond C, Warren-Perry M, Horsley S, Kearney L, Tucker S, Adelman J . Characterization and variation of a human inwardly-rectifying-K-channel gene (KCNJ6): a putative ATP-sensitive K-channel subunit. FEBS Lett 1995; 367: 193–197.
Nakamura A, Hattori M, Sakaki Y . A novel gene isolated from human placenta located in Down syndrome critical region on chromosome 21. DNA Res 1997; 4: 321–324.
Gosset P, Ghezala GA, Korn B, Yaspo ML, Poutska A, Lehrach H, Sinet PM, Creau N . A new inward rectifier potassium channel gene (KCNJ15) localized on chromosome 21 in the Down syndrome chromosome region 1 (DCR1). Genomics 1997; 44: 237–241.
Rao VN, Modi WS, Drabkin HD, Patterson D, O'Brien SJ, Papas TS, Reddy ES . The human erg gene maps to chromosome 21, band q22: relationship to the 8; 21 translocation of acute myelogenous leukemia. Oncogene 1988; 3: 497–500.
Watson DK, McWilliams-Smith MJ, Nunn MF, Duesberg PH, O'Brien SJ, Papas TS . The ets sequence from the transforming gene of avian erythroblastosis virus, E26, has unique domains on human chromosomes 11 and 21: both loci are transcriptionally active. Proc Natl Acad Sci U S A 1985; 82: 7294–7298.
Hyett J, Mosoco G, Nicolaides K . Abnormalities of the heart and great arteries in first trimester chromosomally abnormal fetuses. Am J Med Genet 1997; 69: 207–216.
Schinzel RE . Catalogue of unbalanced chromosome aberrations in man. Berlin: Walter de Gruyter, 1984.
Jackson JF, North ER, Thomas JG . Clinical diagnosis of Down's syndrome. Clin Genet 1976; 9: 483–487.
Preus M . A diagnostic index for Down syndrome. Clin Genet 1977; 12: 47–55.
Mjaatvedt CH, Yamamura H, Wessels A, Ramsdell A, Turner D, Markwald RR . Mechanisms of segmentation, septation, and remodeling of the tubular heart: endocardial cushion fate and cardiac looping. In: Harvey RP, Rosenthal N, editors. Heart development. San Diego: Academic Press, 1999: 159–174.
Gittenberger-de Groot AC, Vrancken Peeters MP, Mentink MM, Gourdie RG, Poelmann RE . Epicardium-derived cells contribute a novel population to the myocardial wall and the atrioventricular cushions. Circ Res 1998; 82: 1043–1052.
Scartezzini P, Egeo A, Colella S, Fumagalli P, Arrigo P, Nizetic D, Taramelli R . Cloning a new human gene from chromosome 21q22.3 encoding a glutamic acid–rich protein expressed in and skeletal muscle. Hum Genet 1997; 99: 387–392.
Vidal-Taboada JM, Lu A, Pique M, Pons G, Gil J, Oliva R . Down syndrome critical region gene 2: expression during mouse development and in human cell lines indicates a function related to cell proliferation. Biochem Biophys Res Commun 2000; 272: 156–163.
Hubert RS, Huo YK, Mitchell S, Yamakawa K, Chen XN, Gadomski C, Noya D, Sun ZG, Kim UJ, Shizuya H, Chen C, Fannin S, de Jong P, Simon M, Korenberg JR . Down syndrome congenital heart disease: physical and transcriptional mapping of the candidate region using BACs and PACs. Am J Hum Genet 1995; 57: A61.
Egeo A, Mazzocco M, Arrigo P, Vidal-Taboada JM, Oliva R, Pirola B, Giglio S, Rasore-Quartino A, Scartezzini P . Identification and characterization of a new human cDNA from chromosome 21q22.3 encoding a basic nuclear protein. Hum Genet 1998; 102: 289–293.
Scott HS, Chen H, Rossier C, Lalioti MD, Antonarakis SE . Isolation of a human gene (HES1) with homology to an Escherichia coli and a zebrafish protein that maps to chromosome 21q22.3. Hum Genet 99: 616–623.
Schmucker D, Clemens JC, Shu H, Worby CA, Xiao J, Muda M, Dixon JE, Zipursky SL . Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell 2000; 101: 671–684.
Agarwala KL, Nakamura S, Tsutsumi Y, Yamakawa K . Down syndrome cell adhesion molecule DSCAM mediates homophilic intercellular adhesion. Brain Res Mol Brain Res 2000; 79: 118–126.
Mjaatvedt CH, Markwald RR . Induction of an epithelial-mesenchymal transition by an in vivo adheron-like complex. Dev Biol 1989; 136: 118–128.
Crossin KL, Hoffman S . Expression of adhesion molecules during the formation and differentiation of the avian endocardial cushion tissue. Dev Biol 1991; 145: 277–286.
Funderburg FM, Markwald RR . Conditioning of native substrates by chondroitin sulfate proteoglycans during cardiac mesenchymal cell migration. J Cell Biol 1986; 103: 2475–2487.
Saffell JL, Williams EJ, Mason IJ, Walsh FS, Doherty P . Expression of a dominant negative FGF receptor inhibits axonal growth and FGF receptor phosphorylation stimulated by CAMs. Neuron 1997; 18: 231–242.
Acknowledgements
This work was supported by NHLBI grants HL50025 (J.R.K. and D.M.K.), HL61033 (J.R.K.), NICHD grant HD17449 (J.R.K.), and Department of Energy grants 92ER61402 and 96ER62294 (J.R.K.). JRK holds the Geri and Richard Brawerman Chair of Molecular Genetics. G.E.L. is supported by a grant from the American Heart Association, WI Affiliate. The authors acknowledge the generous donation of the probes for collagen VI A1 and A2 by Dr. Mon-Li Chu at Thomas Jefferson University, Philadelphia, PA. The authors also thank the DS individuals described herein, together with their families, for their patience and cooperation during the course of this study.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Barlow, G., Chen, XN., Shi, Z. et al. Down syndrome congenital heart disease: A narrowed region and a candidate gene. Genet Med 3, 91–101 (2001). https://doi.org/10.1097/00125817-200103000-00002
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1097/00125817-200103000-00002
Keywords
This article is cited by
-
Identification of novel hypermethylated or hypomethylated CpG sites and genes associated with anthracycline-induced cardiomyopathy
Scientific Reports (2023)
-
Congenital heart defects among Down’s syndrome cases: an updated review from basic research to an emerging diagnostics technology and genetic counselling
Journal of Genetics (2021)
-
A genome-wide search for new imprinted genes in the human placenta identifies DSCAM as the first imprinted gene on chromosome 21
European Journal of Human Genetics (2019)
-
RCAN1 Mutation and Functional Characterization in Children with Sporadic Congenital Heart Disease
Pediatric Cardiology (2018)
-
Down syndrome: from the age of characterization to the era of curative approach
The Nucleus (2017)