Abstract
The IL23R region on chromosome 1 exhibits complex associations with ankylosing spondylitis (AS). We used publicly available epigenomic information and historical genetic association data to identify a putative regulatory element (PRE) in the intergenic region between IL23R and IL12RB2, which includes two single-nucleotide polymorphisms (SNPs) independently associated with AS—rs924080 (P=2 × 10−3) and rs11578380 (P=2 × 10−4). In luciferase reporter assays, this PRE showed silencer activity (P<0.001). Haplotype and conditional analysis of 4230 historical AS cases and 9700 controls revealed a possible AS-associated extended haplotype, including the PRE and risk variants at three SNPs (rs11209026, rs11209032 and rs924080), but excluding the rs11578380 risk variant. However, the rs924080 association was absent after conditioning on the primary association with rs11209032, which, in contrast, was robust to conditioning on all other AS-associated SNPs in this region (P<2 × 10−8). The role of this putative silencer on some IL23R extended haplotypes therefore remains unclear.
Similar content being viewed by others
Introduction
Ankylosing spondylitis (AS) is the archetype of a group of inflammatory disorders, known as spondyloarthropathies, in which inflammation of the spine and sacroiliac joints is prominent.1 It is a polygenic condition for which >100 genetic associations have now been reported.2, 3 Many of the genes implicated in AS are also involved in overlapping clinical conditions, such as psoriasis and inflammatory bowel disease.4, 5 The importance of the interleukin (IL)-23 pathway in AS was initially suggested by the genetic association with the IL-23 receptor (IL23R).6 Subsequently, IL23R expressing cells have been identified at sites of inflammation (enthuses) in mice with SpA.7 The first IL23R association with AS to be described was with rs11209026, a missense variant in the cytoplasmic tail that affects IL-23 R signalling.6, 8, 9 There is also a second independent association with AS in the intergenic region between IL23R and IL12RB2 (encoding the IL12 receptor-specific β chain).3 This association is also apparent in inflammatory bowel disease and Behcet disease.4, 10
Recently, we identified a regulatory enhancer between IL23R and IL12RB2 that is modulated by the AS-associated single-nucleotide polymorphism (SNP) rs11209032, which is in the region of association independent of the primary coding SNP rs11209026.11 Homozygosity for the risk allele ‘A’ at rs11209032 reduces enhancer activity and increases the proportion of IFN-γ-secreting CD4+ (Th1) cells in cases with AS. It is known that genomic regulatory elements can be complex.12 In the present study, we therefore address the possibility that there are other regulatory regions or SNPs associated with this second independent AS association signal. We have used publicly available epigenomic data (that is, ENCODE13 and Roadmap14 projects) to identify additional putative regulatory elements (PREs) overlapping this region of independent genetic association with AS. We have identified a PRE containing two SNPs associated with AS independently of rs11209026, and show that it has silencer activity in luciferase reporter assays. We have performed haplotype analysis and sequential conditional analysis to identify the likely causal variants/haplotypes. Our findings suggest that there may be complex haplotypic regulatory influences in this region, but highlight that the primary AS association is with rs11209032.
Results and discussion
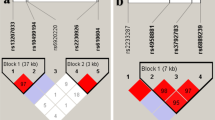
We first analysed the epigenomic landscape of this region in terms of chromatin accessibility and modifications. This identified a PRE (Chr1:67,759,931-67,761,417) located 34.2 kb downstream of IL23R and 11.6 kb upstream of IL12RB2 (Figure 1a) that overlaps a region of independent association previously reported in AS (conditioned on the primary coding SNP rs11209026).3 This region exhibits DNase I hypersensitivity in Th1 and Jurkat cells but not in Th17 cells. Publicly available epigenomic data showed little or no evidence of histone modifications or transcription factor binding,13, 14 but these data are only available for a limited range of conditions of cell activation. We then used luciferase reporter assays to investigate the regulatory activity of this PRE in Jurkat cells transfected with pGL4.23 plasmids containing the minimal promoter with or without relevant variants of the 1.48 kb PRE sequence inserted 5′ of this minimal promoter. Transcriptional activity was compared between the minimal promoter construct alone and the wild-type 1.48 kb PRE sequence (AS-protective ‘C variants’ at both rs924080 and rs11578380), and also for the rs924080 AS-risk ‘T’ variant and rs11578380 AS-risk ‘G’ variant (Figure 1b). The PRE from the wild-type (protective) variant showed reduced reporter activity significantly below the minimal promoter level (P<0.001), suggesting silencer activity. In contrast, the rs924080 AS-risk ‘T’ variant exhibited significantly greater activity than the protective allele (P<0.05), which was not significantly less than that associated with the minimal promoter alone. The AS-risk ‘G’ variant at rs11578380 also significantly increased reporter activity to a similar extent (Figure 1b).
Putative regulatory element containing rs924080 and rs11578380 downstream of IL23R shows silencer activity. (a) Cartoon representation of IL23R and IL12RB2 promoter and putative regulatory element location (Chr1:67,759,931-67,761,417). ENCODE data from UCSC Genome Browser: DNase I hypersensitivity in Th17 cells, Th1 cells and Jurkat cells. (b) The transcriptional activity of rs924080 or rs11578380 compared to the minimal promoter pGL4minP (set to 1) was measured by luciferase reporter assays in Jurkat cells. The values of relative luciferase activity are expressed as mean±s.e.m. of four repeat experiments each carried out in triplicate. Student’s t-test was used. minP, minimal promoter.
To investigate possible regulatory effects arising from multiple SNPs in the IL23R–IL12RB2 intergenic region of association, haplotype analysis was performed using PLINK.15 We identified 10 haplotypes for the AS-associated SNPs rs11209026, rs11209032, rs6677188, rs924080 and rs11578380 (Table 1). The risk alleles at rs11209032 (‘A’ allele) and rs924080 (‘T’ allele) were always co-inherited on haplotypes 2 and 9 (Table 1). The relatively common haplotype 2 was significantly increased in cases (36% AS vs 32% controls, P<2 × 10−21), whereas the rare haplotype 9 occurred at a frequency of only 1% in both cases and controls. The protective haplotype 7 was significantly more frequent in controls (4% AS vs 6% controls, P<3 × 10−18). Together, these data suggest that the risk variants at rs11209026, rs11209032 and rs924080 might have an additive effect since those associated with AS all appear on the same haplotype (haplotype 2). In contrast, the protective variants at rs11578380 and rs6677188 were found on this haplotype, suggesting that these SNPs are unlikely to be involved in any extended haplotypic effect.
To confirm the presence of independent effects from individual SNPs in this putative silencer, we next performed conditional analysis using immunochip data.3 The PRE contains three common SNPs (minor allele frequency >1%), two of which previously showed strong association with AS; rs924080 (P=2 × 10−11) and rs11578380 (P=1 × 10−12).3 The associations at rs924080 (P=2 × 10−3) and rs11578380 (P=2 × 10−4) remained positive after conditioning on rs11209026, confirming an association independent of the coding SNP rs11209026.3 However, this interval of independent association also includes rs11209032, as previously reported.3, 11 After conditioning on rs11209032, the apparent associations at rs924080 and rs11578380 disappeared (Table 2). In contrast, the association with rs11209032 remained positive (P<1 × 10−5) after conditioning on either rs924080 or rs11578380, thereby confirming rs11209032 as the primary association with AS in this intergenic region.
These data could also be relevant to Behcet disease with which rs924080 is strongly associated (P<7 × 10−9).10 Behcet disease is a multisystem inflammatory disorder characterised by symptoms such as uveitis, recurrent oral ulcers, skin lesions and vasculitis; it is also strongly associated with HLA-B*51.16 There are occasional reports of the coexistent AS and Behcet disease, but it remains unclear whether this is incidental or overlaps as part of the extended spectrum of spondyloarthropathies, of which AS is the archetype. Patients with coexistent AS and Behcet disease tend to be positive for both HLA-B*27 (AS-associated) and HLA-B*51, suggesting that it may occur incidentally.17
In conclusion, we have identified a possible silencer downstream of IL23R that includes the AS-associated SNP rs924080, which appears to modulate the functional effects of this regulatory element. We have confirmed the primary association of AS with rs11209032 in this region, but suggest that there could be a possible additional effect from rs924080 in a putative silencer on the same haplotype. Further work is required to determine the potential functional relevance of rs924080 to AS and Behcet disease.
Materials and methods
Identification of a PRE
We used the published data from genome-wide association studies in AS,3 and publicly available data from the ENCODE13 and Roadmap Epigenomics Projects,14 to identify a 1.48 kb PRE downstream of IL23R, which includes two SNPs that are associated with AS—rs924080 and rs11578380. We evaluated DNase I hypersensitivity sites, histone modifications and transcription factor binding-sites previously reported in this region.
Luciferase reporter assay
The relevant 1.48 kb PRE sequence (Chr1:67,759,931-67,761,417) was amplified from genomic DNA. It was cloned into TA cloning kit pCR2.1 vector (Invitrogen, Paisley, UK) and subcloned into pGL4.23(luc2/minP) reporter vector (Promega, Madison, WI, USA) as previously described.11 Point mutations corresponding to genetic variants (T/C) of rs924080 or (G/C) of rs11578380 were introduced (primer sequences available on request) using the QuikChange II XL Site-Directed Mutagenesis Kit (Agilent, Santa Clara, CA, USA). Jurkat cells (Clone E6-1, ATCC TIB-152) were cultured in RPMI supplemented with 10% foetal bovine serum, 100 units per ml penicillin, 100 units per ml streptomycin and 2 mm l-glutamine. A total of 200 000 Jurkat cells per well in 24-well plates were co-transfected with 500 ng of pGL4 construct and 10 ng of pRL-null (Promega) using GeneIn (Amsbio, Abingdon, UK). After 48 h, luciferase activity was measured using the Dual-Luciferase assay reporter system (Promega). Firefly luciferase activity was normalised relative to Renilla luciferase activity for each transfection and calculated as fold increase over pGL4.23(luc2/minP). Four repeat experiments were performed each carried out in triplicate. Student’s t-test was used to determine statistical significance.
Haplotype analysis
All cases and controls were of White British ancestry. IL23R–IL12RB2 haplotypes were estimated for 4230 AS cases and 9700 controls from the IGAS immunochip study3 using PLINK v1.07. The haplotypes for rs11209026, rs11209032, rs6677188, rs924080 and rs11578380 were estimated using the —hap-freq function in PLINK v1.07.15 The χ2-test was used to determine statistical significance.
Conditional analysis
We used only the same subset of cases and controls of White British ancestry from the IGAS immunochip study to evaluate genetic associations with AS.3 Independent effects on genetic susceptibility at the IL23R–IL12RB2 locus were identified by conditional analysis of 4230 AS cases and 9700 matched controls, as previously described.3 Association analysis was performed using the logistic regression function in PLINK v1.07, using five principal components to account for population structure.15
Code availability
PLINK v1.07 was used in this study and can be downloaded from https://www.cog-genomics.org/plink2.
References
Dougados M, van der Linden S, Juhlin R, Huitfeldt B, Amor B, Calin A et al. The European Spondylarthropathy Study Group preliminary criteria for the classification of spondylarthropathy. Arthritis Rheum 1991; 34: 1218–1227.
Ellinghaus D, Jostins L, Spain SL, Cortes A, Bethune J, Han B et al. Analysis of five chronic inflammatory diseases identifies 27 new associations and highlights disease-specific patterns at shared loci. Nat Genet 2016; 48: 510–518.
Cortes A, Hadler J, Pointon JP, Robinson PC, Karaderi T, Leo P et al. Identification of multiple risk variants for ankylosing spondylitis through high-density genotyping of immune-related loci. Nat Genet 2013; 45: 730–740.
Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, Daly MJ et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science 2006; 314: 1461–1463.
Liu Y, Helms C, Liao W, Zaba LC, Duan S, Gardner J et al. A genome-wide association study of psoriasis and psoriatic arthritis identifies new disease loci. PLoS Genet 2008; 4: e1000041.
Burton PR, Clayton DG, Cardon LR, Craddock N, Deloukas P, Duncanson A et al. Association scan of 14,500 nonsynonymous SNPs in four diseases identifies autoimmunity variants. Nat Genet 2007; 39: 1329–1337.
Sherlock JP, Joyce-Shaikh B, Turner SP, Chao C-C, Sathe M, Grein J et al. IL-23 induces spondyloarthropathy by acting on ROR-γt+ CD3+CD4-CD8- entheseal resident T cells. Nat Med 2012; 18: 1069–1076.
Di Meglio P, Di Cesare A, Laggner U, Chu CC, Napolitano L, Villanova F et al. The IL23R R381Q gene variant protects against immune-mediated diseases by impairing IL-23-induced Th17 effector response in humans. PLoS ONE 2011; 6: e17160.
Di Meglio P, Villanova F, Napolitano L, Tosi I, Terranova Barberio M, Mak RK et al. The IL23R A/Gln381 allele promotes IL-23 unresponsiveness in human memory T helper17 cells and impairs Th17 responses in psoriasis patients. J Invest Dermatol 2013; 133: 2381–238.
Remmers EF, Cosan F, Kirino Y, Ombrello MJ, Abaci N, Satorius C et al. Genome-wide association study identifies variants in the MHC class I, IL10, and IL23R-IL12RB2 regions associated with Behcet's disease. Nat Genet 2010; 42: 698–702.
Roberts AR, Vecellio M, Chen L, Ridley A, Cortes A, Knight JC et al. An ankylosing spondylitis-associated genetic variant in the IL23R-IL12RB2 intergenic region modulates enhancer activity and is associated with increased Th1-cell differentiation. Ann Rheum Dis 2016; 75: 2150–2156.
Daniel B, Nagy G, Nagy L . The intriguing complexities of mammalian gene regulation: how to link enhancers to regulated genes. Are we there yet? FEBS Lett 2014; 588: 2379–2391.
ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 2012; 489: 57–74.
Bernstein BE, Stamatoyannopoulos JA, Costello JF, Ren B, Milosavljevic A, Meissner A et al. The NIH Roadmap Epigenomics Mapping Consortium. Nat Biotechnol 2010; 28: 1045–1048.
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81: 559–575.
Takeuchi M, Kastner DL, Remmers EF . The immunogenetics of Behçet's disease: a comprehensive review. J Autoimmun 2015; 64: 137–148.
Küçükşen S, Bağçacı S, Karahan AY, Şahin M, Uğurlu H . Coexistence of Behçet’s disease and ankylosing spondylitis. Eur J Gen Med 2013; 10: 112–114.
Acknowledgements
ARR was funded by Arthritis Research UK (grant 20402), MV by National Institute for Health Research (NIHR) Oxford Comprehensive Biomedical Research Centre (immunity and inflammation theme A93081), AC by a Queensland University of Technology Vice Chancellor’s Research Fellowship, and JCK by the European Research Council under the European Union’s Seventh Framework Programme (FP7/2007-2013)/ERC grant agreement no. 281824, Arthritis Research UK (grant 20773) and the NIHR Oxford Biomedical Research Centre. Additional funding was provided by Arthritis Research UK (grants 19536, 18797 and 20796), the NIHR Thames Valley collaborative research network, NIHR Oxford Musculoskeletal Research Unit and National Ankylosing Spondylitis Society (UK).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Roberts, A., Vecellio, M., Cortes, A. et al. Investigation of a possible extended risk haplotype in the IL23R region associated with ankylosing spondylitis. Genes Immun 18, 105–108 (2017). https://doi.org/10.1038/gene.2017.5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gene.2017.5
This article is cited by
-
Assessment of immunological profile in ankylosing spondylitis patients following a clinical trial with guluronic acid (G2013), as a new NSAID with immunomodulatory properties
Immunologic Research (2019)
-
Genetically determined high activities of the TNF-alpha, IL23/IL17, and NFkB pathways were associated with increased risk of ankylosing spondylitis
BMC Medical Genetics (2018)