Abstract
Single-nucleotide polymorphisms near the interferon lambda 3 (IFNL3) gene predict outcomes to infection and anti-viral treatment in hepatitis C virus (HCV) infection. To identify IFNL3 genotype effects on peripheral blood, we collected phenotype data on 400 patients with genotype 1 chronic hepatitis C (CHC). The IFNL3 responder genotype predicted significantly lower white blood cells (WBCs), as well as lower absolute numbers of monocytes, neutrophils and lymphocytes for both rs8099917 and rs12979860. We sought to define the WBC subsets driving this association using flow cytometry of 67 untreated CHC individuals. Genotype-associated differences were seen in the ratio of CD4CD45RO+ to CD4CD45RO−; CD8CD45RO+ to CD8CD45RO−, NK CD56 dim to bright and monocyte numbers and percentages. Whole blood expression levels of IFNL3, IFNLR1 (interferon lambda receptor 1), IFNLR1-mem (a membrane-associated receptor), IFNLR1-sol (a truncated soluble receptor), MxA and T- and NK (natural killer) cell transcription factors TBX21, GATA3, RORC, FOXP3 and EOMES in two subjects were also determined. CHC patients demonstrated endogenous IFN activation with higher levels of MxA, IFNLR1, IFNLR1-mem and IFNLR1-sol, and IFNL3 genotype-associated differences in transcription factors. Taken together, these data provide evidence of an IFNL3 genotype association with differences in monocyte, T- and NK cell levels in the peripheral blood of patients with CHC. This could underpin genotype associations with spontaneous and treatment-induced HCV clearance and hepatic necroinflammation.
Similar content being viewed by others
Introduction
Hepatitis C virus (HCV) infects over 200 million people.1 Spontaneous clearance of HCV infection is largely affected by variants of the interferon lambda 3 (IFNL3) gene.2 Failure to clear leads to chronic hepatitis C (CHC), which can result in significant complications including liver cirrhosis, hepatocellular carcinoma and death from liver failure, as well as other immune-related phenomena such as cryoglobulinaemia and lymphoma.3
In 2009, three landmark genome-wide association studies (GWAS) identified a set of single-nucleotide polymorphisms (SNPs) in the vicinity of the IFNL3 gene, which were significantly associated with clearance of genotype 1 HCV on conventional therapy.4, 5, 6 Subsequently, this genetic variation has been strongly associated with spontaneous clearance of HCV.2 In 2013, a new polymorphism (ss469415590) between IFNL2 and IFNL3 was identified and found to induce a frameshift mutation, resulting in transient expression of an IFN analogue, IFN lambda 4 (IFNL4), in stimulated human hepatocytes.7 The genotype-dependent production of the protein IFNL4 resulted in altered IFN-sensitive gene (ISG) expression and thus may explain the effects on viral clearance. ss469415590 is in high linkage disequilibrium with rs12979860 but more strongly associated with spontaneous and treatment-induced HCV clearance. We have referred to the SNPs rs12979860 and rs8099917 as IFNL3 SNPs in this paper, although they could also be referred to as being from the gene IFNL4.
IFNL3, a member of the type III IFN family, induces potent innate anti-viral effects against a number of viruses including HCV.8, 9, 10, 11 Its effects are mediated via signalling through the interferon lambda receptor 1 (IFNLR1) complex, whose expression has been confirmed on a variety of cells including lymphocytes.8, 12, 13 There are at least two splice variants of the IFNLR1 receptor chain: a membrane-associated receptor (IFNLR1-mem) and a truncated soluble receptor (IFNLR1-sol), which lacks the transmembrane domain. It has therefore been speculated that the soluble receptor acts as a negative regulator of type III IFNs by binding to the cytokines before cell contact.14 However, soluble receptors can also increase signalling by increasing cytokine half-life15 or potentiating signalling.16
The host immune response is pivotal to a successful outcome at initial infection, during and after development of CHC. A strong virus-specific cytotoxic response, largely mediated by CD4 T helper type 1 (Th1) cells and natural killer (NK) cells, is required to remove infected hepatocytes, secrete cytokines and promote hepatocyte production of ISGs that allow for the inhibition of viral replication.17 In contrast, there is some evidence to suggest that a CD4 Th2-dominant response is associated with HCV treatment failure and viral persistence.18, 19 The anti-viral role of CD8 T cells in CHC is thought to be negligible.20
We hypothesised that IFNL3 genotype may mediate differences in the immunological phenotype in CHC. To test our hypothesis, we initially analysed a large cross-sectional cohort of genotype 1 CHC subjects, then performed a flow cytometric analysis on a cohort of 67 of these. Analysis was also performed on transcription factors, as the master regulators of Th cell and NK differentiation and mediators of the immune response, including: TBX21 (Th1, NK cells), GATA3 (Th2), RORC (Th17), FOXP3 (T-regulatory cells (Tregs)) and EOMES (CD8, NK cells).21 Finally, we also assessed the expression of IFNL3-associated genes (IFNL3, IFNLR1, IFNLR1-mem, IFNLR1-sol and MxA). These data provide evidence of IFNL3 genotype-associated monocyte, T- and NK cell alterations in peripheral blood, which could be due to variation in immune cell trafficking to the infected liver, and may explain the genetic associations with viral clearance, necroinflammation and response to therapy.
Results
Haematological markers correlate with IFNL3 genotype and HCV viral load
The baseline characteristics of the 400 patients according to IFNL3 genotypes are summarised in Table 1. No significant difference between the groups was observed for age or gender. At baseline, a number of highly significant haematological differences between the IFNL3 genotypes were detected (Table 1). The IFNL3 responder genotype groups demonstrated lower baseline white blood cell (WBC) count (rs8099917 P=2.8 × 10−4 and rs12979860 P=2.8 × 10−3), absolute lymphocyte count (ALC) (rs8099917 P=5.0 × 10−3 and rs12979860 P=0.015), absolute neutrophil count (ANC) (rs8099917 P=0.013 and rs12979860 P=0.022) and absolute monocyte count (AMC) (rs8099917 P=6.1 × 10−4 and rs12979860 P=0.021). An association with lower haemoglobin was also observed, but only for rs8099917. There was no significant difference in platelet counts between the IFNL3 genotypes.
HCV viral load measurements were stratified into high (⩾8.5 × 105 IU ml−1) and low (<8.5 × 105IU ml−1) viral load groups. A significantly higher proportion of IFNL3 responder genotypes were observed in the high viral load group compared with the low viral load group (rs8099917: 35% low viral load vs 61% high viral load, P=0.0007 and rs12979860: 16% low viral load vs 50% high viral load, P<0.0001). The high viral load group demonstrated lower WBCs (P=0.034) and ALC (P=0.034) (Table 2).
Flow cytometric deconvolution of leucocytes confirms genotype effect
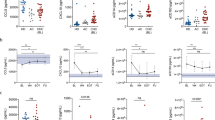
Flow cytometric analysis of peripheral blood mononuclear cells from 67 CHC patients before therapy indicated a reduction in monocytes (P<0.05) in the rs12979860 responder genotype (Figure 1a). Further subsetting demonstrated a reduction in CD56 high NK cells (P<0.05), and a similar trend in CD4+ T cells, as indicated by a reduced CD4/CD8 ratio (P=0.054). Responder genotypes also had a lower proportion of RO+ to RO− in both CD4 and CD8 subsets (P<0.001, P=0.08, respectively).
Flow cytometric analysis of immune cell subsets in CHC by rs12979860 genotype (n=67). (a) Percentage of major immune cell subsets (P<0.05 for monocytes and P=0.05 for CD8s). (b) Ratio of CD56 high to CD56 low cells (P<0.05). (c) Ratio of CD4/CD8 (P⩽0.05). (d) Ratio of CD45RO− to RO+ for CD4 (P<0.001) and CD8 (P=0.08) cells.
Transcription factor expression in peripheral blood in CHC
As CD4CD45RO T and NK cells are thought to mediate viral clearance, we assessed their abundance in peripheral blood by measuring cell subset-specific transcription factors.22, 23 Comparison was made for transcription factors FOXP3, GATA3, RORC and TBX21 between CHC cohort (n=24) and healthy controls (n=22). A significantly higher expression (P=0.04) of circulating FOXP3 cells were detected in CHC cohort compared with healthy controls (Figure 2).
No significant correlation between IFNL3 genotype and transcription factor expression was detected in CHC. As Th1 and NK cells facilitate viral clearance, we hypothesised that the IFNL3 responder genotype would have a Th1, NK-dominant phenotype. To identify relative differences within the lymphocyte population, we used ratios to compare Th1, NK (TBX21) to the other subsets: Th2 (GATA3), Th17 (RORC) and Treg (FOXP3). In addition, as the transcription factors are variably expressed, we used their rank across sample rather than their absolute expression values. For rs8099917, we found a significant association with a TBX21 (Th1/NK)-dominant effect: TBX21/GATA3 (Th1, NK/Th2: P=0.017); TBX21/RORC (Th1, NK/Th17: P=7.4 × 10−3) and TBX21/FOXP3 (Th1, NK:Treg: P=0.036), with the ratio higher for responders in each case. For rs12979860, the same trend was also observed and this was significant for: TBX21/GATA3 (Th1, NK/Th2: P=0.038) and TBX21/RORC (Th1, NK/Th17: P=5.2 × 10−3) (Figure 3). In other works,24 we have confirmed that the expression of these genes is highest in these subsets.
Expression of transcription factor ratios in the peripheral blood of HCV-treated subjects (n=24) analysed for differences in IFNL3 genotype. (a) Ratios show a TBX21 (Th1/NK)-dominant effect associated with the IFNL3 responder genotype (rs8099917 TT and rs12979860 CC). (b) Ratio of TBX21/GATA3 (Th1, NK/Th2) showing significant differences for rs8099917 (P=0.017) and for rs12979860 (P=0.038). (c) Ratio of TBX21/RORC (Th1, NK/Th17) showing significant differences for rs8099917 (P=7.4 × 10−3).
IFNLR1 and MxA mRNAs are increased in peripheral blood in CHC
IFNL3, IFNLR1 and MxA expression was measured in peripheral blood samples from healthy controls (n=22) and HCV-infected untreated subjects (n=24). CHC patients demonstrated significantly higher expression levels of MxA (P=3.0 × 10−6) (Figure 4e), IFNLR1 (P=3.2 × 10−12) (Figure 4b), IFNLR1-mem (P=0.041) (Figure 4c) and IFNLR1-sol (P=3.0 × 10−3) (Figure 4d) compared with healthy controls. However, no difference in IFNL3 mRNA expression was detected between untreated HCV-infected subjects and healthy controls (Figure 4a). We looked for an association between IFNL3 genotype and expression levels of IFNL3, IFNLR1, IFNLR1-sol, IFNLR1-mem and MxA in untreated CHC subjects. There was a trend towards higher baseline expression of all five parameters measured (IFNL3, IFNLR1, MxA, IFNLR1-sol and IFNLR1-mem) compared with those with the IFNL3 responder genotypes (rs809917 TT and rs12979860 CC). However, this only reached statistical significance for IFNLR1-sol (rs809917: P=0.02).
IFNL3, IFNLR1, IFNLR1-mem, IFNLR1-sol and MxA expression by qPCR in peripheral blood from healthy controls (n=22) and untreated HCV-infected patients (n=24). Significantly higher expression of (b) IFNLR1 (P=3.2 × 10−12), (c) IFNLR1-mem (P=0.041) and (d) IFNLR1-sol (P=3.0 × 10−3) and (e) MxA (P=3.0 × 10−6) in the HCV-infected individuals is demonstrated. No difference in (a) IFNL3 expression between controls and HCV subjects in peripheral blood was detected and for rs12979860 (e) at T0 (P=5.2 × 10−3). (c and f) Ratio of TBX21/FOXP3 (Th1:Treg) showing significant differences for rs8099917 (c) at T0 (P=0.036) and T4w (P=0.018).
Discussion
In this study, we sought to define IFNL3 genotype effects on peripheral blood immune cells to improve our understanding of the basis IFNL3 genotype-associated differences in viral clearance. We demonstrate, for the first time, significantly lower baseline total white cell, neutrophil, lymphocyte and monocyte counts for people with IFNL3 responder genotypes (for both rs8099917 and rs12979860) and an association between higher pre-treatment viral load and lower white cell and lymphocyte counts. From flow cytometric analysis, responder genotypes had a lower CD56 high/dim ratio, lower CD45RO+/− ratio and fewer monocytes. The IFN-sensitive genes MxA and IFNLR (sol- and membrane-bound isoforms) were higher in CHC compared with that in controls, but IFNL3 was not. For responders for all genes there was a trend for higher expression.
Further, in patients with IFNL3 responder genotype, transcription factor analysis revealed evidence for a Th1/NK-dominant state in peripheral blood. Taken together, these results suggest that in individuals with the IFNL3 responder genotype there may be increased lymphocyte redistribution to the liver and secondary lymphoid organs, resulting in increased priming and activation of adaptive immune cells. On treatment with exogenous IFNA, the Th1/NK-dominant response increased immune cell activation and immune cell residency in the liver would favour rapid viral clearance and may explain IFNL3 genotype-associated associations with rapid virological response and sustained virological response.
Lymphopenia is associated with viral infections and others have shown that this is related to redistribution of lymphocytes to secondary lymphoid organs25 and increased lymphocyte tissue residency time.26 This lymphocyte redistribution is mediated, in part, by endogenous type I IFNs in viral infection.27 Cirrhosis is also associated with haematologic abnormalities including varying degrees of cytopenias. However, in neither instance has this been observed to relate to type III IFN genotype. Moreover, in cirrhosis, thrombocytopenia is the most commonly detected abnormality, with low WBCs and anaemia tending to develop later in the disease course.28 In our study, we saw no consistent association of platelet counts or haemoglobin with IFNL3 genotype, suggesting a targeted effect on immune cell activation and trafficking. Thus, our data suggest an IFNL3 genotype-specific altered immune state and blood profile, likely triggered by chronic viral infection and endogenous IFNs.
In support of this contention, we demonstrate evidence of endogenous activation of the peripheral blood compartment by the IFN system in CHC. Significantly higher levels of MxA have been described previously,29 and here we additionally show that expression of IFNLR1, IFNLR1-mem and IFNLR1-sol are also elevated in infected patients compared with healthy controls. Interestingly, we did not observe higher expression levels for IFNL3 mRNA in HCV-infected subjects, compared with controls. IFNL3 production is produced by rare immune cell subsets, including BDCA3 dendritic cells and plasmacytoid dendritic cells, as we and others have previously demonstrated.13, 30 It is therefore possible that differences in production of IFNL3 between healthy controls and CHC subjects may be only detectable by analysing these immune cell subsets.
In relation to IFNL3 genotype, we observed higher ISG expression in the peripheral blood of patients with the responder genotype, using MxA as the candidate ISG. Patients with the IFNL3 responder genotype have been shown to express more IFNL3, yet demonstrate lower hepatic ISG expression compared with non-responders at baseline.31, 32, 33 Taken together, these data suggest that IFNL3 may act predominantly on immune cells to facilitate HCV clearance in these individuals.
Since the publication of genome-wide association studies in 2009 identifying SNPs in the vicinity of IFNL3 associated with response to treatment in CHC, there has been an intense research effort to determine the molecular basis for the genotype effect. A number of significant advances have been reported, including replicated associations of the favourable IFNL3 genotype with lower hepatic ISG expression,31, 32, 33 increased hepatic necroinflammation,34, 35, 36, 37 higher baseline viral load4, 38, 39 and increased rates of rapid virological response/early virological response.40, 41, 42, 43 However, the basis for these associations remains largely conjectural. An altered host immune state linked to the IFNL3 polymorphism may result in a Th1/NK-dominant response to HCV infection. Although Th1/NK cytokines favour viral clearance, they are likely to also have a role in mediating hepatocellular damage, if clearance is not achieved. Among the Th1/NK cytokines interleukin-2, IFN-γ and tumour necrosis factor-α have been shown to mediate tissue injury,17 whereas high levels have been associated with lymphopenia, particularly T-cell lymphopenia.27 Those with the responder genotype have fewer effectors T cells, fewer NK CD56 bright than dim and lower monocyte counts, which may be due to increased trafficking or redistribution of these cells to the liver and secondary lymphoid tissues, resulting in increased priming and activation of adaptive immune cells. Following treatment with exogenous IFNA, the milieu in IFNL3 responder genotype individuals renders them primed for viral clearance. This is particularly the case as IFNL3 has the ability to modulate Tregs and enhance the adaptive cellular response through induction of Th1-biased responses.44
Our study has several limitations, including the small number of patients with detailed kinetic and transcription factor analysis. Further, the observed genotype associations with transcription factor ratios may be driven by immune cells other than those from which their expression was expected. The Th subset (Th1, Th2, Treg and Th17) and NK findings should ultimately be confirmed with cytokine and flow cytometric analysis of peripheral blood. Ideally, but perhaps unrealistically, paired liver biopsies would be required to validate our hypothesis of altered cell trafficking to the liver in IFNL3 genotypes, but the recent report by Honda et al.45 suggests that this is indeed the case. Interestingly, a recent genome-wide association study subanalysis from the Individualized Dosing Eficacy vs. Flat Dosing to Assess Optimal Pegylated Interferon Therapy (IDEAL) study,41 performed to detect SNPs associated with cytopenias during treatment, failed to detect any association with IFNL3 genotypes.46 There is some evidence that this cohort had less advanced liver disease compared with our patients, including milder fibrosis, higher platelet counts and younger age (summarised in Supplementary Table 2). In addition, there were a number of exclusion criteria in the IDEAL study (including low pre-treatment ANC, platelet and haemoglobin), which did not apply to our patient population, and it is known that lower WBCs and anaemia tend to develop later in the disease course.28 Thus, the IFNL3 genotype effect we observed may be cohort-dependent and requires replication.
In summary, we observed an altered pre-treatment immune state in the peripheral blood of patients with genotype 1 CHC, with reduced numbers of WBC, ANC, ALC and ANC and a Th1/NK bias in the IFNL3 responder genotypes. Compared with controls, CHC subjects demonstrated evidence of endogenous IFN activation with higher expression levels of MxA, IFNLR1, IFNLR1-mem and IFNLR1-sol, but not IFNL3. These novel and highly significant associations with IFNL3 genotype strengthen support for an immune cell-mediated foundation for the molecular basis of this genotype effect.
Materials and methods
Study cohort
The three study cohorts consisted of Caucasian subjects with genotype 1 HCV infection (summarised in Figure 5). Briefly, baseline haematological data were collected on an initial cohort of 400 subjects (Cohort 1). HCV viral load measurements were also available for 171 of these subjects. From Cohort 1, 67 subjects (Cohort 2) were studied with a flow cytometric panel. Expression of a panel of genes was undertaken for 24 of these subjects (Cohort 3) using PAXgene Blood RNA tubes (Qiagen, Valencia, CA, USA). In addition, PAXgene Blood RNA tubes were collected from 22 age-matched healthy Caucasian controls for comparison.
Ethics statement
Ethical approval was obtained from the Human Research Ethics Committees of the Sydney West Area Health Service and the University of Sydney. All subjects gave written informed consent (HREC2002/12/4.9(1564)).
Flow cytometry
Venous blood was collected in EDTA and peripheral blood mononuclear cells isolated on Ficoll-Paque Plus (VWR International, Brisbane, QLD, Australia), washed in phosphate-buffered saline and cryopreserved in RPMI-1640 medium (Life Technologies, Carlsbad, CA, USA) containing 2 mm glutamine, 10% heat-inactivated foetal bovine serum (Fisher Biotec, Wembley, WA, Australia), 10% dimethyl sulphoxide and 50 U ml−1 penicillin and 50 μg ml−1 streptomycin. Peripheral blood mononuclear cells were thawed, washed in RPMI with 2% foetal bovine serum and incubated for 30 min in RPMI with 2% foetal bovine serum, 10 mm HEPES, 1 mm magnesium chloride and 100 U ml−1 DNase I (Roche, Sydney, NSW, Australia). Antibodies used were: mAb to CD19-BV421 (HIB19), CD3-PE (UCHT1) and CD4-BV570 (RPA-T4) from BioLegend (San Diego, CA, USA); CD14-PerCP (MφP9), CD56-PECy-7 (NCAM16.2), CD8-BV650 (RPA-T8), CD45RO-APC-H7 (UCHL1), T-Bet-Alexa Fluor 647 (4B10) and corresponding isotype control (IgG1) from BD Biosciences (San Jose, CA, USA); Eomes-FITC (WD1928) and corresponding isotype control (IgG1) from ebioscience (San Diego, CA, USA). Cells were blocked with mouse IgG (33 μg ml−1; Life Technologies) and stained for all extracellular antigens except CD14 in Brilliant Stain Buffer (BD Horizon, San Jose, CA, USA). Cells were fixed, permeabilised, blocked in mouse serum and stained for Eomes and T-bet (or corresponding isotype controls) and CD14 using the Foxp3 Staining Buffer Set (ebioscience) according to the manufacturer’s instructions. Cells were analysed on a Fortessa (BD Biosciences) using the FlowJo software (Tree star Inc., Ashland, OR, USA).
IFNL3 genotyping and gene expression by qPCR
All healthy controls and HCV-infected subjects (summarised in Table 1) were genotyped for the rs8099917 and rs12979860 SNPs by methods reported previously.13, 47
Total RNA was extracted from whole blood in PAXgene tubes using the PAXgene Blood RNA Kit (Qiagen, Hilden, Germany). The RNeasy Kit (Qiagen, Valencia, CA, USA) was used for immune cell pellets. cDNA was prepared using Superscript III, RNaseOUT, OligodT12–18 primer and random primers (Life Technologies) in a Mastercycler gradient 5331 (Eppendorf AG, Hamburg, Germany). Reverse transcription was performed at 50 °C for 45 min.
Gene expression was measured by quantitative PCR (qPCR) using custom-designed primers (Sigma-Aldrich, St Louis, MO, USA) specific for IFNL3 (forward: 5′-CCCAAAAAAGGAGTCCCCTG-3′ and reverse: 5′-GGTTGCATGACTGGCGGA-3′). Specificity for IFNL3 was confirmed by sequencing of the PCR pro(methods published previously13). In addition, primers for IFNLR1 were designed (forward: 5′-CTAAGCCCACCTGCTTCTTG-3′; reverse: 5′-GTCAGTTCCTTTTGGGGACA-3′). These primers detected both splice forms of IFNLR1: the membrane-associated and soluble forms. Primers for the membrane-associated (INLR1-mem, forward: 5′-CTAAGCCCACCTGCTTCTTG-3′; reverse: 5′-TGTCCCCAAAAGGAACTGAC-3′) and soluble receptor (INLR1-sol, forward: 5′-CTAAGCCCACCTGCTTCTTG-3′; reverse: 5′-TGTCCCCAAAAGGAACTGAC-3′) were also designed.13 MxA (forward: 5′-GCCGGCTGTGGATATGCTA-3′; reverse: 5′-TTTATCGAAACATCTGTGAAAGCAA-3′) was selected as the candidate ISG given published associations with HCV treatment outcomes.48 GAPDH primers (forward: 5′-TCCACCACCCTGTTGCTGTA-3′; reverse: 5′-ACCACAGTCCAGCCATCAC-3′) were used as the housekeeping gene. Amplification was measured using Power SYBR Green PCR Master Mix (Life Technologies). Gel electrophoresis was used to confirm the absence of gDNA products from the PCR reactions. Expression was measured using CT values, normalised to that of GAPDH (ΔCT=CT (GAPDH)−CT (target) and then expressed as . CT values were <30, and all amplifications were carried out in duplicate. Transcription factor expression was assessed using Taqman Gene Expression assays from Applied Biosystems (Carlsbad, CA, USA).
Statistics
Baseline and 4-week data for various demographic, haematological, biochemical and virological characteristics are expressed as mean±s.d. The difference between IFNL3 genotypes was assessed by χ2 test or t-test where appropriate. qPCR data were transformed for normality (log(Y × 106). Pearson's R coefficient was used to determine the correlation between samples. Transcription factor expression levels across the samples were ranked, and the ratio of the ranks was compared. A two-sided P-value <0.05 was considered significant.
References
Messina JP, Humphreys I, Flaxman A, Brown A, Cooke GS, Pybus OG et al. Global distribution and prevalence of hepatitis C virus genotypes. Hepatology 2015; 61: 77–87.
Thomas DL, Thio CL, Martin MP, Qi Y, Ge D, O'Huigin C et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature 2009; 461: 798–801.
Hoofnagle JH . Course and outcome of hepatitis C. Hepatology 2002; 36 (Suppl 1): S21–S29.
Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature 2009; 461: 399–401.
Suppiah V, Moldovan M, Ahlenstiel G, Berg T, Weltman M, Abate ML et al. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat Genet 2009; 41: 1100–1104.
Tanaka Y, Nishida N, Sugiyama M, Kurosaki M, Matsuura K, Sakamoto N et al. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet 2009; 41: 1105–1109.
Prokunina-Olsson L, Muchmore B, Tang W, Pfeiffer RM, Park H, Dickensheets H et al. A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nat Genet 2013; 45: 164–171.
Ank N, West H, Bartholdy C, Eriksson K, Thomsen AR, Paludan SR . Lambda interferon (IFN-lambda), a type III IFN, is induced by viruses and IFNs and displays potent antiviral activity against select virus infections in vivo. J Virol 2006; 80: 4501–4509.
Coccia EM, Severa M, Giacomini E, Monneron D, Remoli ME, Julkunen I et al. Viral infection and Toll-like receptor agonists induce a differential expression of type I and lambda interferons in human plasmacytoid and monocyte-derived dendritic cells. Eur J Immunol 2004; 34: 796–805.
Marcello T, Grakoui A, Barba-Spaeth G, Machlin ES, Kotenko SV, MacDonald MR et al. Interferons alpha and lambda inhibit hepatitis C virus replication with distinct signal transduction and gene regulation kinetics. Gastroenterology 2006; 131: 1887–1898.
Robek MD, Boyd BS, Chisari FV . Lambda interferon inhibits hepatitis B and C virus replication. J Virol 2005; 79: 3851–3854.
Siebler J, Wirtz S, Weigmann B, Atreya I, Schmitt E, Kreft A et al. IL-28 A is a key regulator of T-cell-mediated liver injury via the T-box transcription factor T-bet. Gastroenterology 2007; 132: 358–371.
O’Connor KS, Ahlenstiel G, Suppiah V, Schibeci S, Ong A, Leung R et al. IFNL3 mediates interaction between innate immune cells: implications for hepatitis C virus pathogenesis. Innate Immunity 2013; 20: 598–605.
Witte K, Gruetz G, Volk HD, Looman AC, Asadullah K, Sterry W et al. Despite IFN-lambda receptor expression, blood immune cells, but not keratinocytes or melanocytes, have an impaired response to type III interferons: implications for therapeutic applications of these cytokines. Genes Immunity 2009; 10: 702–714.
Peters M, Jacobs S, Ehlers M, Vollmer P, Mullberg J, Wolf E et al. The function of the soluble interleukin 6 (IL-6) receptor in vivo: sensitization of human soluble IL-6 receptor transgenic mice towards IL-6 and prolongation of the plasma half-life of IL-6. J Exp Med 1996; 183: 1399–1406.
Bergamaschi C, Bear J, Rosati M, Beach RK, Alicea C, Sowder R et al. Circulating IL-15 exists as heterodimeric complex with soluble IL-15Ralpha in human and mouse serum. Blood 2012; 120: e1–e8.
Nelson DR, Lau JY . Pathogenesis of hepatocellular damage in chronic hepatitis C virus infection. Clin Liver Dis 1997; 1: 515–528, v.
Ueyama M, Nakagawa M, Sakamoto N, Onozuka I, Funaoka Y, Watanabe T et al. Serum interleukin-6 levels correlate with resistance to treatment of chronic hepatitis C infection with pegylated-interferon-alpha2b plus ribavirin. Antiviral Ther 2011; 16: 1081–1091.
Guzman-Fulgencio M, Jimenez JL, Berenguer J, Fernandez-Rodriguez A, Lopez JC, Cosin J et al. Plasma IL-6 and IL-9 predict the failure of interferon-alpha plus ribavirin therapy in HIV/HCV-coinfected patients. J Antimicrobial Chemother 2012; 67: 1238–1245.
Rehermann B . Pathogenesis of chronic viral hepatitis: differential roles of T cells and NK cells. Nat Med 2013; 19: 859–868.
Luckheeram RV, Zhou R, Verma AD, Xia B . CD4(+)T cells: differentiation and functions. Clin Dev Immunol 2012; 2012: 925135.
Parnell GP, Gatt PN, Krupa M, Nickles D, McKay FC, Schibeci SD et al. The autoimmune disease-associated transcription factors EOMES and TBX21 are dysregulated in multiple sclerosis and define a molecular subtype of disease. Clin Immunol 2014; 151: 16–24.
Edstrom M, Mellergard J, Mjosberg J, Jenmalm M, Vrethem M, Press R et al. Transcriptional characteristics of CD4+ T cells in multiple sclerosis: relative lack of suppressive populations in blood. Mult Scler 2011; 17: 57–66.
Shahijanian F, Parnell GP, McKay FC, Gatt PN, Shojoei M, O'Connor KS et al. The CYP27B1 variant associated with an increased risk of autoimmune disease is underexpressed in tolerizing dendritic cells. Hum Mol Genet 2013; 23: 1425–1434.
Baenziger S, Heikenwalder M, Johansen P, Schlaepfer E, Hofer U, Miller RC et al. Triggering TLR7 in mice induces immune activation and lymphoid system disruption, resembling HIV-mediated pathology. Blood 2009; 113: 377–388.
Gunzer M, Riemann H, Basoglu Y, Hillmer A, Weishaupt C, Balkow S et al. Systemic administration of a TLR7 ligand leads to transient immune incompetence due to peripheral-blood leukocyte depletion. Blood 2005; 106: 2424–2432.
Kamphuis E, Junt T, Waibler Z, Forster R, Kalinke U . Type I interferons directly regulate lymphocyte recirculation and cause transient blood lymphopenia. Blood 2006; 108: 3253–3261.
Qamar AA, Grace ND, Groszmann RJ, Garcia-Tsao G, Bosch J, Burroughs AK et al. Incidence, prevalence, and clinical significance of abnormal hematologic indices in compensated cirrhosis. Clin Gastroenterol Hepatol 2009; 7: 689–695.
Mihm S, Frese M, Meier V, Wietzke-Braun P, Scharf JG, Bartenschlager R et al. Interferon type I gene expression in chronic hepatitis C. Lab Invest 2004; 84: 1148–1159.
Yoshio S, Kanto T, Kuroda S, Matsubara T, Higashitani K, Kakita N et al. Human blood dendritic cell antigen 3 (BDCA3)(+) dendritic cells are a potent producer of interferon-lambda in response to hepatitis C virus. Hepatology 2013; 57: 1705–1715.
Dill MT, Duong FH, Vogt JE, Bibert S, Bochud PY, Terracciano L et al. Interferon-induced gene expression is a stronger predictor of treatment response than IL28B genotype in patients with hepatitis C. Gastroenterology 2011; 140: 1021–1031.
Honda M, Sakai A, Yamashita T, Nakamoto Y, Mizukoshi E, Sakai Y et al. Hepatic ISG expression is associated with genetic variation in interleukin 28B and the outcome of IFN therapy for chronic hepatitis C. Gastroenterology 2010; 139: 499–509.
Urban TJ, Thompson AJ, Bradrick SS, Fellay J, Schuppan D, Cronin KD et al. IL28B genotype is associated with differential expression of intrahepatic interferon-stimulated genes in patients with chronic hepatitis C. Hepatology 2010; 52: 1888–1896.
Bochud PY, Bibert S, Kutalik Z, Patin E, Guergnon J, Nalpas B et al. IL28B alleles associated with poor hepatitis C virus (HCV) clearance protect against inflammation and fibrosis in patients infected with non-1 HCV genotypes. Hepatology 2012; 55: 384–394.
Agundez JA, Garcia-Martin E, Maestro ML, Cuenca F, Martinez C, Ortega L et al. Relation of IL28B gene polymorphism with biochemical and histological features in hepatitis C virus-induced liver disease. PLoS One 2012; 7: e37998.
Thompson AJ, Clark PJ, Zhu M, Zhu Q, Ge D, Sulkowski MS et al. Genome side association study identified IL28B polymorphism to be associated with baseline ALT and hepatic necro-inflammatory activity in chronic hepatitis C patients enrolled in the IDEAL study. Hepatology 2010; 52 (Suppl): 1220A–1221A.
Jouvin-Marche E, Macek Jilkova Z, Thelu MA, Marche H, Fugier E, Van Campenhout N et al. Lymphocytes degranulation in liver in hepatitis C virus carriers is associated with IFNL4 polymorphisms and ALT levels. J Infect Dis 2014; 209: 1907–1915.
Bochud PY, Bibert S, Negro F, Haagmans B, Soulier A, Ferrari C et al. IL28B polymorphisms predict reduction of HCV RNA from the first day of therapy in chronic hepatitis C. J Hepatol 2011; 55: 980–988.
McCarthy JJ, Li JH, Thompson A, Suchindran S, Lao XQ, Patel K et al. Replicated association between an IL28B gene variant and a sustained response to pegylated interferon and ribavirin. Gastroenterology 2010; 138: 2307–2314.
Kurosaki M, Tanaka Y, Nishida N, Sakamoto N, Enomoto N, Honda M et al. Pre-treatment prediction of response to pegylated-interferon plus ribavirin for chronic hepatitis C using genetic polymorphism in IL28B and viral factors. J Hepatol 2011; 54: 439–448.
McHutchison JG, Lawitz EJ, Shiffman ML, Muir AJ, Galler GW, McCone J et al. Peginterferon alfa-2b or alfa-2a with ribavirin for treatment of hepatitis C infection. N Engl J Med 2009; 361: 580–593.
Naggie S, Osinusi A, Katsounas A, Lempicki R, Herrmann E, Thompson AJ et al. Dysregulation of innate immunity in hepatitis C virus genotype 1 IL28B-unfavorable genotype patients: impaired viral kinetics and therapeutic response. Hepatology 2012; 56: 444–454.
Par G, Szereday L, Berki T, Palinkas L, Halasz M, Miseta A et al. Increased baseline proinflammatory cytokine production in chronic hepatitis C patients with rapid virological response to peginterferon plus ribavirin. PLoS One 2013; 8: e67770.
Morrow MP, Pankhong P, Laddy DJ, Schoenly KA, Yan J, Cisper N et al. Comparative ability of IL-12 and IL-28B to regulate Treg populations and enhance adaptive cellular immunity. Blood 2009; 113: 5868–5877.
Honda M, Shirasaki T, Shimakami T, Sakai A, Horii R, Arai K et al. Hepatic interferon-stimulated genes are differentially regulated in the liver of chronic hepatitis C patients with different interleukin 28B genotypes. Hepatology 2013; 59: 828–838.
Thompson AJ, Clark PJ, Singh A, Ge D, Fellay J, Zhu M et al. Genome-wide association study of interferon-related cytopenia in chronic hepatitis C patients. J Hepatol 2012; 56: 313–319.
Suppiah V, Moldovan M, Ahlenstiel G, Berg T, Weltman M, Abate ML et al. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat Genet 2009; 41: 1100–1104.
McGilvray I, Feld JJ, Chen L, Pattullo V, Guindi M, Fischer S et al. Hepatic cell-type specific gene expression better predicts HCV treatment outcome than IL28B genotype. Gastroenterology 2012; 142: 1122–1131.
Acknowledgements
We thank all patients for their valuable participation in this study. We would like to acknowledge Elizabeth Snape and Vincenzo Fragomeli for assistance with sample collections. This project was supported, in part, by the Robert W Storr Bequest to the Sydney Medical Foundation, University of Sydney; National Health and Medical Research Council of Australia (NHMRC) Program Grant No. 1053206; and a University of Sydney Bridging Support Grant.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on Genes and Immunity website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
O'Connor, K., Read, S., Wang, M. et al. IFNL3/4 genotype is associated with altered immune cell populations in peripheral blood in chronic hepatitis C infection. Genes Immun 17, 328–334 (2016). https://doi.org/10.1038/gene.2016.27
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gene.2016.27
This article is cited by
-
The presence of interferon affects the progression of non-alcoholic fatty liver disease
Genes & Immunity (2022)