Abstract
Tractable experimental model that accounts for inter-tumor molecular heterogeneity is a key element of anti-cancer drug development. Hepatocellular carcinoma is known to exhibit highly heterogeneous molecular aberrations across the tumors, including somatic genetic and epigenetic alterations. Previous studies showed that molecular tumor subtypes determined by transcriptome, as a comprehensive functional readout, are reproducibly observed across global patient populations irrespective of geographic and etiological variations. Here we demonstrate that transcriptomic hepatocellular carcinoma subtypes, S1 and S2, determined by our previous transcriptome meta-analysis of multiple clinical hepatocellular carcinoma cohorts, are presented in a panel of hepatoma cell lines widely used by the research community. Interestingly, cell line that resembles gene expression pattern of S3 subtype, representing less aggressive tumors, was not identified in the panel. MYC pathway-activated S2-like cell lines showed higher sensitivity to a small molecule BET bromodomain inhibitor, (+)-JQ1, which has anti-MYC activity. These results support the use of hepatoma cell lines as models to evaluate molecular subtype-specific drug response, which is expected to lead to development of tailored, precision care of the patients with hepatocellular carcinoma.
Similar content being viewed by others
Introduction
Experimental models are key components of anti-cancer drug development and an urgent unmet need for hepatocellular carcinoma (HCC), which still lacks substantially effective medical therapy.1 Extensive genomic analysis of HCC tumors has clarified significant inter- and intra-tumor molecular heterogeneity, which likely obscures therapeutic effect of molecular targeted agents. Potentially druggable somatic DNA mutations are generally rare (prevalence <5%), which makes patient enrollment in clinical trials more challenging.2 A recent study of patient-derived HCC cell lines demonstrated selective killing only in subclones that harbor the relatively rare somatic DNA aberrations such as FGF19 gene amplification.3
Transcriptome is an alternative measure, which is assumed to be functional readouts of various types of molecular aberrations such as mutations and epigenetic changes. Our previous transcriptome meta-analysis revealed three subtypes, namely S1, S2 and S3, which are observed in larger fractions of patients/tumors (prevalence>20%) associated with specific molecular pathway dysregulation and patient prognosis.1, 4, 5 The subtypes are associated with clinical characteristics such as serum α-fetoprotein (AFP) levels, and also with histological intra-tumor heterogeneity, suggesting their clinical relevance.5 It is empirically known that some of the transcriptomic subtypes are present in human hepatoma cell lines, with which subtype-specific in vitro and in vivo anti-cancer drug response has been demonstrated.6, 7, 8 In parallel, clinical features associated with the subtypes such as AFP (a marker of the S2 subtype) have been recognized as possible predictive biomarkers of therapeutic response to molecular targeted agents as shown in recent clinical trials.9 These findings collectively suggest that hepatoma cell lines can be used as tractable and clinically relevant experimental models of the HCC subtypes for assessment of molecular targeted anti-cancer agents. In this study, we have systematically evaluated the HCC subtypes and their association with therapeutic response in a panel of frequently and widely used hepatoma cell lines.
Materials and methods
Transcriptomic subtyping of hepatoma cell lines
Pre-normalized genome-wide transcriptome and DNA mutation data sets of 25 hepatoma cell lines (Table 1) were downloaded from the Cancer Cell Line Encyclopedia (CCLE) website (https://portals.broadinstitute.org/ccle/home). Pre-normalized genome-wide transcriptome data set of 374 human HCC tissues was downloaded from The Cancer Genome Atlas (TCGA) website (https://gdc.cancer.gov). In the CCLE hepatoma data set, transcriptome profile of PLC/PRF/5 cell line duplicated in an alternative name (Alexander) was excluded. Hepatoblastoma-derived cell lines, Hep G2 and its derivative C3A,10 and undifferentiated hepatoma cell lines, HLE and HLF, were included in the analysis given their wide use in the HCC research community. Transcriptomic subtyping of the cell lines was performed based on the HCC subtype maker gene signatures by using the Nearest Template Prediction algorithm as previously reported.4, 11 Non-negative matrix factorization (NMF)-based consensus clustering was used to explore optimal number of classes in the global transcriptome data set after excluding less variable probes based on a coefficient of variation (CV) cutoff of 0.5.12
Cell lines
SNU-182, SNU-387, SNU-475, SNU-449, Hep G2, Hep 3B2.1-7, Huh-7 and THLE-5B cells (American Type Culture Collection or Riken Bioresource, Japan) were grown in DMEM supplemented with 10% heat-inactivated fetal bovine serum, 100 U ml−1 penicillin (Gibco, Gaithersburg, MD, USA) and 100 μg ml−1 streptomycin (Gibco) at 37 °C in a 5% CO2 atmosphere.
HCC subtyping with NanoString assay
Total RNA was extracted from the cell lines using RNeasy kit (Qiagen, Germantown, MD, USA), and 200 ng total RNA was subjected to the Elements HCC subtyping assay (NanoString, Seattle, WA, USA) as previously reported.5 Raw transcript count data were normalized by scaling with geometric mean of built-in normalization genes. HCC subtype determination was similarly performed by the Nearest Template Prediction algorithm.
In vitro anti-HCC drug treatment
A total of 3000–5000 cells were plated in each well in 96-well plates according to growth rate of each cell line, and when 50% confluence was reached ~24 h later, treatment with (+)-JQ1 (Selleckchem, Houston, TX, USA) at varying concentrations was started and continued for 48 h in triplicates.
Cell viability analysis
Cell viability was assessed using an MTS Cell Proliferation Assay (Promega, Madison, WI, USA). A volume of 22 μl of the MTS assay solution was added to each well that contained 110 μl of media. The plates were then incubated at 37 °C in a 5% CO2 atmosphere for 4 h. The absorbance was then read at 490 nm. Averaged absorbance from empty wells was subtracted from all wells as background signal, and relative cell viability was calculated as a percentage of the adjusted absorbance compared to respective DMSO-treated control wells.
Statistical analysis
Proportion of categorical data was assessed by Fisher’s exact test. Two-tailed P-value <0.05 was regarded as statistically significant. All analyses were performed using R statistical language (www.r-project.org).
Results
Transcriptomic subtyping of hepatoma cell lines
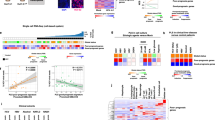
Our previous transcriptome meta-analysis involving 603 clinical HCC tissues revealed the following three subtypes: S1 subtype characterized by activation of stromal gene/pathway such as transforming growth factor β (TGFβ), S2 subtype with overexpression of stemness markers such as EPCAM and AFP, and S3 subtype enriched with more differentiated tumors and accumulation of CTNNB1 exon 3 mutations associated with induction of liver-specific Wnt pathway target, GLUL.13 It has been reported that hepatoma cell lines are classified into two groups by unsupervised clustering of global transcriptome profiles, which is correlated with AFP expression status.14, 15 AFP-high and -low cell lines were empirically known to exhibit transcriptome profiles similar to S2 and S1 subtypes, respectively.4 Consistent with these observations, non-negative matrix factorization consensus clustering of the genome-wide transcriptome profiles of 25 hepatoma cell lines from the CCLE data set revealed that optimal classification is indeed achieved when two subclasses are assumed (Figure 1a). In the surgically resected clinical HCC tissues (n=374), median expression levels of each HCC subtype marker genes in each sample (S1: 238 genes, S2: 115 genes and S3: 266 genes) depicted relative abundance of each subtype marker gene expression in human HCC tissues (Figure 1b).
Transcriptomic hepatocellular carcinoma (HCC) subtypes and DNA mutations in 25 hepatoma cell lines. (a) Optimal number of unsupervised subclasses in the global transcriptome profiles of the 25 hepatoma cell lines from the Cancer Cell Line Encyclopedia (CCLE) data set was determined by cophenetic coefficient from non-negative matrix factorization (NMF) consensus clustering for each assumed number of subclasses, ranging from 2 to 5. (b) Median expression levels of the HCC subtype marker genes across the samples are plotted for each subtype in 374 clinical HCC tissues (left) and 25 hepatoma cell lines (right). Boxes represent 75th and 25th percentile, horizontal line is the median, and whiskers mark lowest and highest values. Outliers outside 1.5 × of inter-quartile range are shown as open circles. (c) GLUL expression level in the 25 hepatoma cell lines. Arrow head indicates a cell line with CTNNB1 exon 3 mutation.
In the hepatoma cell lines, the relative expression levels of the S1 and S2 subtype marker genes were maintained, whereas the S3 subtype marker genes were not expressed at the levels in tissues, suggesting that the hepatoma cell lines generally lack characteristic of the S3 subtype. In fact, the CTNNB1 exon 3 mutations prevalent in the S3 subtype, was observed only in one cell lines (SNU-398) with relatively low expression of the target gene GLUL (Figures 1c and 2). These results collectively support that the hepatoma cell lines are classified into either the S1 or S2 subtype, and there is no cell line corresponding to the S3 subtype. In contrast, TP53 gene mutations were more frequently observed (12 out of 22 cell lines with mutation data, 55%) as seen in clinical tissues, and HIF1A was the most frequently affected gene by mutations (68%). Prevalence of these mutations was similar between the HCC subtypes for TP53 (P=0.67, Fisher’s exact test), HIF1A (P=0.38), TERT promoter (P=1.0) and rest of the genes recurrently affected in human HCC tissues (P=0.83).2
Transcriptomic hepatocellular carcinoma (HCC) subtypes and DNA mutations in 25 hepatoma cell lines. HCC subtypes were determined using genome-wide transcriptome data by the Nearest Template Prediction (NTP) algorithm as previously described.4 Expression pattern of the HCC subtype marker genes is shown in the heatmap.
Transcriptomic subtyping of hepatoma cells by an FDA-approved assay
We have implemented a reduced version of the HCC subtyping gene signature (30-gene signature) in an FDA-approved diagnostic assay platform (NanoString) for clinical application in our previous study.5 The HCC subtype determination based on the genome-wide transcriptome data (Figure 2) was verified by using the NanoString assay for six representative hepatoma cell lines (SNU-465, SNU-449, SNU-387, Huh-7, Hep 3B and Hep G2) together with the non-tumorigenic SV40-immortalized hepatocyte line, THLE-5B.16 The relative expression of subtype S1 and S2 marker genes as well as low expression of S3 marker genes was verified in the hepatoma cells, and a similar pattern was observed in THLE-5B cells (Figure 3a). This result suggests that the lower expression of the S3 marker genes compared to HCC tissues is a common feature in established cell lines irrespective of origin, that is, malignant or non-malignant cells. The HCC subtype determination was also confirmed in the 30-gene HCC subtyping assay, supporting validity of the assay (Figure 3b).
Transcriptomic hepatocellular carcinoma (HCC) subtypes determined by the NanoString assay. (a) Median expression levels of the HCC subtype marker genes are plotted for each subtype in seven cell lines. Red dots indicate expression in THLE-5B cells. Boxes represent 75th and 25th percentile, horizontal line is the median, and whiskers mark lowest and highest values. Outliers outside 1.5 × of inter-quartile range are shown as open circles. (b) Expression pattern of the HCC subtype marker genes in the seven cell lines is shown in the heatmap.
HCC subtypes are associated with response to molecular targeted agents
In our previous study, we could successfully demonstrate in vitro and in vivo subtype-specific drug response by targeting fibroblast growth factor receptor pathway, one of the molecular hallmarks specifically activated in the S2 subtype.8 We sought to extend this examination to another pathway implicated in HCC subtypes. MYC pathway is known to be activated in the S2 subtype.4 Direct targeting of this pathway has been technically challenging, but BET bromodomain inhibition by a small molecule, (+)-JQ1 has been show to elicit anti-MYC pathway activity.17, 18 To evaluate whether the subtype-dependent response to MYC pathway inhibition can be monitored in hepatoma cells, SNU-387 and SNU-182 (S1-like cells) as well as Huh-7 and Hep G2 (S2-like cells) were treated with (+)-JQ1. In the CCLE data set, expression of the target genes, BRD2, BRD3 or BRD4, at naive state was confirmed in the S2-like cell lines, which are already known to exhibit MYC pathway activation (Figure 4a).4 As expected, the S2-like cell lines showed higher sensitivity to (+)-JQ1 as indicated by lower half maximal inhibitory concentration (IC50) values (Figure 4b), further supporting the use of hepatoma cell lines as experimental models to assess molecular subtype-specific anti-HCC drug response.
Transcriptomic hepatocellular carcinoma (HCC) subtype-dependent drug response in hepatoma cell lines. (a) Expression of BRD2, BRD3 and BRD4 genes, encoding targets of a BET bromodomain inhibitor, (+)-JQ1, in the S1-like (SNU-387 and SNU-182) and S2-like (Huh-7 and Hep G2) hepatoma cell lines in the CCLE data set. (b) Relative cell viability compared to DMSO-treated controls after 48 h treatment with (+)-JQ1 in a series of drug concentrations (range: 0.01–13.6 μM).
Discussion
This study provides comprehensive reference information for a panel of widely used and commercially available hepatoma cell lines as in vitro models of human HCC molecular subtypes. Alternative strategies such as patient-derived xenograft (PDX) could enable personalized molecular evaluation for each patient.19 However, the low success rate and long time required to establish viable lines limit their flexible use. In contrast, despite the well-recognized limitations such as lacked tissue architecture and crosstalk with other cell types, cancer cell lines remain one of the major experimental models for anti-cancer drug discovery and assessment as a tractable system easily amenable to high-throughput and high-content molecular analysis.20, 21
Modeling of specific disease contexts such as clinical/molecular tumor subtypes is generally attempted by genetic engineering in experimental systems. However, it is practically infeasible to reconstitute the full spectrum of complex genetic and epigenetic molecular aberrations generally observed in non-hereditary solid cancers. Our study highlights an advantage of using existing hepatoma cell lines that inherently exhibit and recapitulate naturally occurring transcriptomic dysregulation observed in two of the three consensus human HCC molecular subtypes, which represent more advanced and aggressive tumors and therefore should be prioritized for therapeutic development. In fact, multiple preclinical studies and clinical trials support this concept. Dasatinib (Src/Abl inhibitor) was more effective in the S1-like cells.6 S2-like cells were more susceptible to epidermal growth factor receptor (EGFR) and insulin-like growth factor 1 receptor (IGF1R) inhibition.22, 23 BGJ398 (pan-fibroblast growth factor receptor inhibitor) elicited in vivo anti-tumor effect in xenograft of the S2-like cells.8 Epitope-optimized genetic vaccines targeting AFP (S2 marker)-induced specific CD8 T-cell-mediated killing of HCC cells in mice.24 Galunisertib, which targets the TGFβ pathway, a hallmark of the S1 subtype, is currently under evaluation in a phase 2 trial (ClinicalTrials.gov, NCT01246986).25 Tivantinib (MET inhibitor),26 ramucirumab (VEGFR2 inhibitor, showed better response in AFP-high patients)9 and GC33 (humanized monoclonal antibody against glypican 3 [GPC3])27, 28 were tested in clinical trials and could be more effective in the S2 subtype.
In our analysis of 25 hepatoma cell lines and one immortalized hepatocyte line, we could not find any cell line with the features of the S3 subtype. This may indicate that HCC cells in S3 tumors, clinically more differentiated and associated with better prognosis, are not capable of survival and proliferation in culture without support from the physiological liver tissue architecture. It is important to note that there is certain diversity in the characteristic molecular features for each subtype between the cell lines. Therefore, presence or absence of specific features (for example, AFP secretion status), which are potentially relevant to mechanism of action for agents to be tested, must be confirmed before picking cell lines for evaluation. In addition, differences in the sources and/or passages could alter the status of transcriptomic subtypes and other genetic aberrations, which ideally should be verified prior to the assessment. In summary, noting these limitations, hepatoma cell lines with designated HCC subtypes represent an invaluable resource as experimental models that retain the intra/inter-tumor/patient heterogeneity for anti-cancer drug assessment and development.
References
Goossens N, Sun X, Hoshida Y . Molecular classification of hepatocellular carcinoma: potential therapeutic implications. Hepat Oncol 2015; 2: 371–379.
Shibata T, Aburatani H . Exploration of liver cancer genomes. Nat Rev Gastroenterol Hepatol 2014; 11: 340–349.
Gao Q, Wang ZC, Duan M, Lin YH, Zhou XY, Worthley DL et al. Cell culture system for analysis of genetic heterogeneity within hepatocellular carcinomas and response to pharmacologic agents. Gastroenterology 2017; 152: 232–242 e4.
Hoshida Y, Nijman SM, Kobayashi M, Chan JA, Brunet JP, Chiang DY et al. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res 2009; 69: 7385–7392.
Tan PS, Nakagawa S, Goossens N, Venkatesh A, Huang T, Ward SC et al. Clinicopathological indices to predict hepatocellular carcinoma molecular classification. Liver Int 2016; 36: 108–118.
Finn RS, Aleshin A, Dering J, Yang P, Ginther C, Desai A et al. Molecular subtype and response to dasatinib, an Src/Abl small molecule kinase inhibitor, in hepatocellular carcinoma cell lines in vitro. Hepatology 2013; 57: 1838–1846.
Deshmukh M, Hoshida Y . Genomic profiling of cell lines for personalized targeted therapy for hepatocellular carcinoma. Hepatology 2013; 58: 2207.
Schmidt B, Wei L, DePeralta DK, Hoshida Y, Tan PS, Sun X et al. Molecular subclasses of hepatocellular carcinoma predict sensitivity to fibroblast growth factor receptor inhibition. Int J Cancer 2016; 138: 1494–1505.
Zhu AX, Park JO, Ryoo BY, Yen CJ, Poon R, Pastorelli D et al. Ramucirumab versus placebo as second-line treatment in patients with advanced hepatocellular carcinoma following first-line therapy with sorafenib (REACH): a randomised, double-blind, multicentre, phase 3 trial. Lancet Oncol 2015; 16: 859–870.
Lopez-Terrada D, Cheung SW, Finegold MJ, Knowles BB . Hep G2 is a hepatoblastoma-derived cell line. Human Pathol 2009; 40: 1512–1515.
Hoshida Y . Nearest Template Prediction: a single-sample-based flexible class prediction with confidence assessment. PLoS ONE 2010; 5: e15543.
Brunet JP, Tamayo P, Golub TR, Mesirov JP . Metagenes and molecular pattern discovery using matrix factorization. Proc Natl Acad Sci USA 2004; 101: 4164–4169.
Rebouissou S, Franconi A, Calderaro J, Letouze E, Imbeaud S, Pilati C et al. Genotype-phenotype correlation of CTNNB1 mutations reveals different ss-catenin activity associated with liver tumor progression. Hepatology 2016; 64: 2047–2061.
Kawai HF, Kaneko S, Honda M, Shirota Y, Kobayashi K . Alpha-fetoprotein-producing hepatoma cell lines share common expression profiles of genes in various categories demonstrated by cDNA microarray analysis. Hepatology 2001; 33: 676–691.
Lee JS, Thorgeirsson SS . Functional and genomic implications of global gene expression profiles in cell lines from human hepatocellular cancer. Hepatology 2002; 35: 1134–1143.
Mace K, Aguilar F, Wang JS, Vautravers P, Gomez-Lechon M, Gonzalez FJ et al. Aflatoxin B1-induced DNA adduct formation and p53 mutations in CYP450-expressing human liver cell lines. Carcinogenesis 1997; 18: 1291–1297.
Aird F, Kandela I, Mantis C. Reproducibility Project: Cancer B. Replication Study: BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Elife 2017; 6: pii: e21253.
Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 2011; 146: 904–917.
Eirew P, Steif A, Khattra J, Ha G, Yap D, Farahani H et al. Dynamics of genomic clones in breast cancer patient xenografts at single-cell resolution. Nature 2015; 518: 422–426.
Boehm JS, Golub TR . An ecosystem of cancer cell line factories to support a cancer dependency map. Nat Rev Genet 2015; 16: 373–374.
Bouhaddou M, DiStefano MS, Riesel EA, Carrasco E, Holzapfel HY, Jones DC et al. Drug response consistency in CCLE and CGP. Nature 2016; 540: E9–E10.
Fuchs BC, Fujii T, Dorfman JD, Goodwin JM, Zhu AX, Lanuti M et al. Epithelial-to-mesenchymal transition and integrin-linked kinase mediate sensitivity to epidermal growth factor receptor inhibition in human hepatoma cells. Cancer Res 2008; 68: 2391–2399.
Zhao H, Desai V, Wang J, Epstein DM, Miglarese M, Buck E . Epithelial-mesenchymal transition predicts sensitivity to the dual IGF-1R/IR inhibitor OSI-906 in hepatocellular carcinoma cell lines. Mol Cancer Ther 2012; 11: 503–513.
Hong Y, Peng Y, Guo ZS, Guevara-Patino J, Pang J, Butterfield LH et al. Epitope-optimized alpha-fetoprotein genetic vaccines prevent carcinogen-induced murine autochthonous hepatocellular carcinoma. Hepatology 2014; 59: 1448–1458.
Giannelli G, Mikulits W, Dooley S, Fabregat I, Moustakas A, ten Dijke P et al. The rationale for targeting TGF-beta in chronic liver diseases. Eur J Clin Invest 2016; 46: 349–361.
Santoro A, Rimassa L, Borbath I, Daniele B, Salvagni S, Van Laethem JL et al. Tivantinib for second-line treatment of advanced hepatocellular carcinoma: a randomised, placebo-controlled phase 2 study. Lancet Oncol 2013; 14: 55–63.
Zhu AX, Gold PJ, El-Khoueiry AB, Abrams TA, Morikawa H, Ohishi N et al. First-in-man phase I study of GC33, a novel recombinant humanized antibody against glypican-3, in patients with advanced hepatocellular carcinoma. Clin Cancer Res 2013; 19: 920–928.
Ikeda M, Ohkawa S, Okusaka T, Mitsunaga S, Kobayashi S, Morizane C et al. Japanese phase I study of GC33, a humanized antibody against glypican-3 for advanced hepatocellular carcinoma. Cancer Sci 2014; 105: 455–462.
Knowles BB, Howe CC, Aden DP . Human hepatocellular carcinoma cell lines secrete the major plasma proteins and hepatitis B surface antigen. Science 1980; 209: 497–499.
Huh N, Utakoji T . Production of HBs-antigen by two new human hepatoma cell lines and its enhancement by dexamethasone. Gan 1981; 72: 178–179.
Nagamori S, Fujise K, Hasumura S, Homma S, Sujino H, Matsuura T et al. [Protein secretion of human cultured liver cells]. Hum Cell 1988; 1: 382–390.
Hasumura S, Sujino H, Nagamori S, Kameda H . [Establishment and characterization of a human hepatocellular carcinoma cell line JHH-4]. Hum Cell 1988; 1: 98–100.
Homma S, Nagamori S, Fujise K, Hasumura S, Sujino H, Matsuura T et al. [Establishment and characterization of a human hepatocellular carcinoma cell line JHH-7 producing alpha-fetoprotein and carcinoembryonic antigen—changes in secretion of AFP and CEA from JHH-7 cells after heat treatment]. Hum Cell 1990; 3: 152–157.
Alexander JJ, Bey EM, Geddes EW, Lecatsas G . Establishment of a continuously growing cell line from primary carcinoma of the liver. S Afr Med J 1976; 50: 2124–2128.
Park JG, Lee JH, Kang MS, Park KJ, Jeon YM, Lee HJ et al. Characterization of cell lines established from human hepatocellular carcinoma. Int J Cancer 1995; 62: 276–282.
Lee JH, Ku JL, Park YJ, Lee KU, Kim WH, Park JG . Establishment and characterization of four human hepatocellular carcinoma cell lines containing hepatitis B virus DNA. World J Gastroenterol 1999; 5: 289–295.
Dor I, Namba M, Sato J . Establishment and some biological characteristics of human hepatoma cell lines. Gan 1975; 66: 385–392.
Acknowledgements
This work was supported by Japan Society for the Promotion of Science, Program for advancing strategic international networks to accelerate the circulation of talented researchers (to TH), Uehara Memorial Foundation (to SN) and NIH/NIDDK DK099558, European Union ERC-2014-AdG-671231 HEPCIR, Irma T Hirschl Trust, US Department of Defense W81XWH-16-1-0363 (to YH).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Hirschfield, H., Bian, C., Higashi, T. et al. In vitro modeling of hepatocellular carcinoma molecular subtypes for anti-cancer drug assessment. Exp Mol Med 50, e419 (2018). https://doi.org/10.1038/emm.2017.164
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/emm.2017.164
This article is cited by
-
Osimertinib is a dual inhibitor of hepatocellular carcinoma and angiogenesis in an EGFR-independent manner, and synergizes with venetoclax
Journal of Cancer Research and Clinical Oncology (2023)
-
Molecular classification of hepatocellular carcinoma: prognostic importance and clinical applications
Journal of Cancer Research and Clinical Oncology (2022)
-
Systematic Analysis of Cytostatic TGF-Beta Response in Mesenchymal-Like Hepatocellular Carcinoma Cell Lines
Journal of Gastrointestinal Cancer (2021)
-
lncRNA HOTAIR overexpression induced downregulation of c-Met signaling promotes hybrid epithelial/mesenchymal phenotype in hepatocellular carcinoma cells
Cell Communication and Signaling (2020)
-
Harnessing big ‘omics’ data and AI for drug discovery in hepatocellular carcinoma
Nature Reviews Gastroenterology & Hepatology (2020)