Abstract
DNA methylation may regulate gene expression by restricting the access of transcription factors. We have previously demonstrated that GATA-1 regulates the transcription of the CCR3 gene by dynamically interacting with both positively and negatively acting GATA elements of high affinity binding in the proximal promoter region including exon 1. Exon 1 has three CpG sites, two of which are positioned at the negatively acting GATA elements. We hypothesized that the methylation of these two CpGs sites might preclude GATA-1 binding to the negatively acting GATA elements and, as a result, increase the availability of GATA-1 to the positively acting GATA element, thereby contributing to an increase in GATA-1-mediated transcription of the gene. To this end, we determined the methylation of the three CpG sites by bisulfate pyrosequencing in peripheral blood eosinophils, cord blood (CB)-derived eosinophils, PBMCs, and cell lines that vary in CCR3 mRNA expression. Our results demonstrated that methylation of CpG sites at the negatively acting GATA elements severely reduced GATA-1 binding and augmented transcription activity in vitro. In agreement, methylation of these CpG sites positively correlated with CCR3 mRNA expression in the primary cells and cell lines examined. Interestingly, methylation patterns of these three CpG sites in CB-derived eosinophils mostly resembled those in peripheral blood eosinophils. These results suggest that methylation of CpG sites at the GATA elements in the regulatory regions fine-tunes CCR3 transcription.
Similar content being viewed by others
Introduction
CCR3 is constitutively expressed at high levels in eosinophils with 16,000-60,000 receptors per cell; it serves as the primary chemokine receptor responsible for eosinophil recruitment to inflamed tissues (Daugherty et al., 1996; Kitaura et al., 1996; Ponath et al., 1996). CCR3 is also expressed on prominent allergic inflammatory cells, including Th2 helper cells (Sallusto et al., 1997) and mast cells (Forsythe and Befus, 2003). The restricted expression of CCR3 leads to a notion that it plays an integral role in the pathogenesis of allergic diseases including asthma, allergic dermatitis, and allergic rhinitis. Furthermore, as airway epithelial cells express functional CCR3, this chemokine receptor is postulated to play other roles beyond trafficking of the migratory cells, such as airway remodeling (Stellato et al., 2001; Beck et al., 2006). Thus, the exact role of CCR3, which is expressed on a variety of cell types, in the development of allergic symptoms remains to be established. The interest in CCR3 as a therapeutic target led to the development of a number of small molecule antagonists, some of which have been studied in clinical trials against eosinophil-associated diseases (Willems and Ijzerman, 2010). The pathological role of eosinophils has been challenged with the findings that anti-interleukin-5 (IL-5) antibody for anti-eosinophil therapy had little effect on clinical parameters seen in mild-to-moderate asthma (Leckie et al., 2000; Flood-Page et al., 2003). However, this therapy has recently been shown to be effective for the treatment of a small group of patients with eosinophilic asthma (Haldar et al., 2009; Nair et al., 2009). In this sense, it has been suggested that targeting both CCR3 and IL-5 might be a more effective means in reducing eosinophil recruitment and function (Rådinger and Lötvall, 2009).
CCR3 expression is mainly regulated at the transcriptional level. The critical sequence that is responsible for transcriptional regulation of CCR3 has been mapped to exon 1 and its flanking sequences (Zimmerman et al., 2000; Scotet et al., 2001; Vijh et al., 2002; Zimmerman et al., 2005). This sequence includes binding elements for GATA-1, acute-myeloid leukemia-1 (AML-1), PU.1, and the CCAAT enhancer binding protein, most of which are also known to participate in the regulation of eosinophil development/differentiation in a combinatorial manner (Zhang et al., 1997; Nerlov and Graf, 1998; Hirasawa et al., 2002; Iwama et al., 2002; McNagny and Graf, 2002), suggesting an intimate relationship between CCR3 expression and eosinophil development. Among these, GATA-1 is considered the most critical transcription factor for both eosinophil development and eosinophil-specific gene expression. GATA-1 binds to a GATA site within the murine GATA-1 promoter with a high affinity, and removal of the binding site selectively abolishes the eosinophil lineage (Yu et al., 2002). GATA-1 transactivates eosinophil-specific genes, including major basic protein (MBP), Charcot-Leyden crystal protein, and eosinophil-derived neurotoxin, by binding to functional GATA elements in their promoters (Dyer and Rosenberg, 2000; Du et al., 2002; Qiu et al., 2009). Involvement of GATA-1 in CCR3 transcription has been demonstrated (Zimmerman et al., 2005) and subsequently leads to postulation of a double-GATA element as a key regulatory element of GATA-1-mediated transcription of eosinophil-specific genes, including human CCR3, interleukin-5 receptor alpha, MBP and GATA-1 genes (Rothenberg and Hogan, 2006). We have recently analyzed GATA-1-mediated transcription of CCR3 at the molecular level (Kim et al., 2010). Of five GATA elements in exon 1 of CCR3, the first GATA element is solely responsible for GATA-1-mediated transctivation. The second and third GATA elements have minimal effects on reporter activity, with low binding affinity for GATA-1. In contrast, the fourth and fifth GATA elements play an inhibitory role in transactivation, with a high affinity binding for GATA-1 comparable to that of the first GATA element. Thus, it appears that the positively acting GATA element exerts its effect in conjunction with the negatively acting GATA elements for GATA-1-mediated transcriptional control of CCR3.
In mammals, the majority of DNA methylation occurs symmetrically on the cytosine residues on both strands of the CpG dinucleotide. DNA methylation can directly inhibit promoter function by sterically hindering the binding of transcription factors and indirectly affecting the chromatin state through the recruitment of methyl-CpG-binding proteins, such as MeCP2 (Klose and Bird, 2006). CpG-rich promoters demonstrate very low levels of CpG methylation at the transcription start sites, while CpG-poor promoters exhibit markedly higher levels of CpG methylation (Zhang et al., 2009). CpG methylation is an essential feature of development, as it reprograms the transcriptional activation and inactivation of genes during development (Okano et al., 1999; Li, 2002; Reik, 2007). Methylation of CpGs serves as dynamic epigenetic marks that undergo extensive change during cellular differentiation. Promoters with low CpG density are associated with developmentally regulated genes, while promoters with high CpG density are associated with constitutively expressed genes (Meissner et al., 2008). It has recently been demonstrated that DNA methylation plays a direct role in lineage restriction during hematopoiesis process, and alterations in expression of lineage-specific genes accompany graded changes in DNA methylation within those genes in a differentiation stage-specific manner during lineage specification (Broske et al., 2009; Trowbridge et al., 2009).
CCR3 expression is subject to epigenetic regulation. Treatment with histone deacetylase inhibitors results in induction of CCR3 mRNA in myeloid cell lines (Tiffany et al., 1998; Ishihara et al., 2007; Kim et al., 2010). However, whether DNA methylation is involved in expression of the gene has not been reported. Close examination shows that exon 1 of CCR3 includes three CpG sites, two of which are located in the regions immediately flanking the fourth GATA element and within the fifth GATA element, respectively. As the fourth and fifth GATA elements may act as sinkers of GATA-1 due to their high affinity binding for GATA-1 against the first GATA element that is responsible for transactivation, methylation of these sites can affect GATA-1-mediated CCR3 transcription. These observations prompted us to investigate whether these CpG sites could be methylated in primary eosinophils and a variety of cell lines that greatly vary in CCR3 mRNA expression and whether methylation of these sites influences GATA-1 binding and the resulting CCR3 transcription.
Results
CpG sites at GATA elements in the regulatory region of CCR3 gene
The CCR3 gene is located on chromosome 3p21 and consists of at least four exons (Vijh et al., 2002). This gene does not have a CpG island throughout its entire sequence of promoter, exons, and introns, as judged on the basis of its size, GC content, and CG dinucleotide frequency (Zhao and Han, 2009), thus indicating a CpG-poor promoter or regulatory region. The most critical regulatory sequences for CCR3 gene transcription reside in exon 1 of 161 base pairs in length (Vijh et al., 2002), which includes five GATA sites, two AML-1 sites, and a CREB site (Figure 1). The fourth and the fifth GATA sites in exon 1 are claimed to constitute a double-GATA site as a key element that dictates GATA-1-mediated transcription of many eosinophil-specific genes (Rothenberg and Hogan, 2006). However, our previous study using a reporter plasmid assay in K562 cells demonstrated that the first GATA site is solely responsible for GATA-1-mediated transactivation of CCR3, while the fourth and fifth GATA sites play inhibitory roles in CCR3 transcription, with high affinity binding for GATA-1 comparable to that of the first GATA element (Kim et al., 2010).The differential contributions of these GATA sites to CCR3 transcription were almost exactly duplicated in A549 cells (Supplemental Data Figure S1), and GATA-1 bound to sequences in exon 1 of CCR3 genes, as analyzed by ChIP assay (Supplemental Data Figure S2). Close examination revealed that there are three CpG sites in exon 1. One is located within the CREB binding element and the other two reside in the regions immediately flanking the fourth GATA site and within fifth GATA site (Figure 1). The negative regulation of the fourth and fifth GATA sites in CCR3 transcription led to the hypothesis that methylation of these two sites might limit GATA-1 binding and redirect available GATA-1 to the positive GATA element, resulting in an increase in GATA-1-mediated transcription of CCR3.
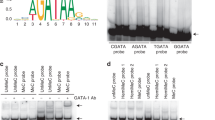
The sequences of exon 1 of CCR3 and the probes for EMSA. (A) Exon 1 is 161 base pairs in length. Numbers on the right are designated based on the longest exon (Vijh et al., 2002). Open and closed boxes represent CREB and GATA elements, respectively. AML-1 elements are indicated by a double line. Asterisks indicate CpG sites, one of which is located within the CREB element, and the other two are located in regions flanking and within GATA elements, respectively. (B) Three probe sequences for EMSA are indicated. Probe 1 includes two CpG sites, while probe 2 and probe 3 include a single CpG site.
Methylation of three CpG sites in exon 1 of CCR3 in peripheral blood eosinophils, CB-derived eosinophils, and a panel of cell lines
The methylation of the CpG sites in exon 1 was quantitatively determined by pyrosequencing following bisulfite treatment of genomic DNA from peripheral blood eosinophils (Figure 2 and Table 1). Each CpG site displayed a characteristic methylation pattern with slight variation among donors: methylation of the CpG site at the CREB site was low, the 4th GATA site was intermediately-to-highly methylated, and the 5th GATA site was highly methylated. We also determined the methylation of these CpG sites in human cord blood (CB)-derived eosinophils, which are shown to express significant levels of CCR3 mRNA (Lamkhioued et al., 2003; Kang et al., 2005). To this end, CD34+ cells were purified to 95% homogeneity by MACS and were cultured in the presence of a cytokine cocktail for 24 days. CD34+ cells gradually disappeared, while MBP-positive cells appeared as early as day 4 (data not shown), steadily increased, and attained a greater than 90% proportionality at day 18 (Figure 3A). Immunofluorescence analysis showed that nearly all cultured cells had a high density of MBP molecules in their cytoplasmic granules at day 24 (Figure 3B), confirming that CB cells are a suitable in vitro model of eosinophils. CB-derived eosinophils had low CpG methylation at the CREB site (2-4%), intermediateto-high methylation at the fourth GATA element (71-84%), and high methylation at the fifth GATA element (82-90%) (Figure 2). Thus, this methylation pattern was strikingly similar to what were observed in peripheral blood eosinophils (Figure 2). Methylation of the the CpG at the CREB site in PBMC was at intermediate level (47-58%), while the fourth and fifth GATA sites were highly methylated, as in peripheral blood eosinophils and CB-derived eosinophils (Table 1). We further analyzed the methylation statuses of a panel of cell lines (Table 1). The CpG site at the CREB element was hypermethylated in five cell lines, including U937, Jurkat, NCI-H292, A549, and 293T, while the CpG site was hypomethylated in two myeloid cell lines, including K562 and HL-60 cells. The CpG methylation at the fourth and fifth GATA elements was high in U937, Jurkat, and NCI-H292 cells, intermediate in A549 and 293T cells, and low in K562 and HL-60 cells. Thus, none of these cell lines were similar to peripheral blood eosinophils or CB-derived eosinophils in the methylation patterns of these CpG sites. It was of interest to note that HL-60 cells, which are converted to eosinophilic cells with induced expression of surface CCR3 in the presence of histone deacetylase inhibitors (Tiffany et al., 1998; Ishihara et al., 2007), had entirely different methylation patterns at the three sites from peripheral blood eosinophils and CB-derived eosinophils (Table 1).
DNA methylation of CpG sites in exon 1 analyzed by bisulfite pyrosequencing. The panels represent methylation status of CpG sites in exon 1 of the CCR3 gene in peripheral blood eosinophils and CB-derived eosinophils. The positions of CpG sites are indicated on the upper histograms, and the methylation levels are presented on the top of each histogram. In parenthesis, the CpG sites at +31, +103, and +118 are located in the CREB, fourth GATA, and fifth GATA elements, respectively.
Production of CB-derived eosinophils. (A) CD34+ cells were cultured with a cytokine cocktail for the days indicated, as described in Materials and Methods. Expressions of CD34 and MBP proteins were analyzed by FACS. The results represent the mean ± SEM of three to seven independent experiments. (B) MBP expression of developing eosinophils at day 0 and 24 was visualized with a fluorescence microscope. DAPI and MBP stains are shown in blue and green, respectively.
Correlation of CCR3 mRNA expression with CpG methylation
To explore correlation between CCR3 mRNA expression and methylation of these CpG sites quantitative real-time PCR of CCR3 mRNA was carried out whose levels were expressed as ΔCT (CCR3 CT -GAPDH CT) following real-time PCR. The results showed that CCR3 mRNA was expressed at the highest level in peripheral blood eosinophils, followed by CB-derived eosinophils and PBMCs. U937 and Jurkat cells expressed CCR3 mRNA at lower levels than did peripheral blood eosinophils and CB-derived eosinophils but at higher levels than three epithelial cell lines. Interestingly, two myeloid cell lines, K562 and HL-60, expressed negligible levels of CCR3 mRNA in an undifferentiated state. The ΔCT values for CCR3 mRNA in all the cell types tested correlated with the levels of methylation at both the fourth and the fifth GATA elements: Pearson correlation coefficients (r) were -0.543 (P = 0.030) and -0.694 (P = 0.003) at the fourth and the fifth GATA elements, respectively (Figure 4), suggesting an association of methylation at these CpG sites with CCR3 mRNA expression.
Correlation of methylation of CpG sites at GATA elements with CCR3 mRNA level. The levels of methylation in the fourth (A) and fifth (B) GATA sites were determined using bisulfite pyrosequencing, and CCR3 mRNA expression was measured by real-time PCR, as described in Methods. The correlation between the level of CCR3 mRNA expression and the degree of methylation on CpG was analyzed using Pearson correlation analysis.
Binding of GATA-1 to GATA elements with methylated CpG sites
To evaluate the effect of methylation on GATA-1 binding, EMSA was carried out with nuclear extracts of K562 cells endogenously expressing a high level of GATA-1 (Du et al., 2002). The fourth and fifth GATA elements in the exon 1 sequence contain CpGs, 5'-tatc cg-3' for fourth GATA and 5'-tatc g-3' for fifth GATA, where underlined text denotes the GATA elements and bold text denotes CpG sites (Figure 1). We synthesized oligonucleotides in which the dC bases at +103 and +118 were converted to deoxymethylcytosine, with the complementary antisense oligonucleotides also methylated at the corresponding dCs to those in the sense strands. Methylated probe 1 bound to GATA-1 more weakly than the unmethylated counterpart (lane 4 versus lane 1 in Figure 5A). The specificity of GATA-1 binding was confirmed by formation of a supershift upon incubation with anti-GATA-1, even though the methylated probe bound very weakly (lanes 7 versus 9 in Figure 5A). The difference in the binding affinity of these two oligonucleotides was evident by the extent of binding with increasing amounts of nuclear extracts (Figure 5B). Probe 2, which contained a CpG site at a region immediately flanking the fourth GATA element, showed slightly reduced binding upon methylation (lane 1 versus lane 2 in Figure 5C), while probe 3, which contained a CpG site within the fifth GATA element, exhibited nearly no binding upon methylation (lane 3 versus lane 4 in Figure 5C). Therefore, the inability of the methylated CpG within the fifth GATA element to bind GATA-1 greatly contributed to the significantly reduced binding of methylated probe 1 that contained most of the sequences of both probe 2 and probe 3. In addition, the severe impairment of GATA-1 binding to the methylated fifth GATA element is in line with more significant and obvious correlation of the methylated fifth GATA element with CCR3 mRNA expression than the methylated fourth GATA element, as seen in Figure 4. Competition experiments revealed that methylated CpG sites had an approximately 2.5-fold weaker affinity for GATA-1 than did the unmethylated sites (Figure 5D), as measured by densitiometry analysis. These results suggest that GATA elements with methylated CpGs have much lower binding affinity for GATA-1 in vitro compared to that of those with unmethylated CpGs. Next, we tested the differential in vivo occupancy of GATA-1 to the unmethylated and methylated GATA elements in K562 cells and EoL-1 cells in which the GATA elements were hypomethylated and hypermethylated, respectively (Table 1). However, we observed the GATA-1 occupancy regardless of cell types (data not shown). Lack of the differential binding seems to be due to the fact that the current ChIP assay cannot discern, if any, differential binding of GATA-1 to these GATA sites, probably because the fourth and fifth GATA elements are positioned at only about 100 base pair away from the first GATA element that shows a high affinity binding for GATA-1, as seen in previous results (Kim et al., 2010).
EMSA using probes with methylated and unmethylated CpG sites. (A) Unmethylated (U) and methylated probe 1 (M), which includes the two CpG sites as depicted in Figure 1, were incubated with nuclear extracts (4 mg) from K562 cells in the presence or absence of indicated competitors (50 ×). Specificity of GATA-1 binding was confirmed by preincubation with specific anti-GATA-1 (S) and control antibody (C). Arrow head and arrow indicate protein-DNA complex and supershift, respectively. (B) To examine binding efficiency, increasing amounts (0.5, 1, 2, and 4 µg) of nuclear extracts (NE) were incubated with unmethylated and methylated probe 1. (C) To determine which methylated CpG contributed to the inefficient binding of methylated probe 1 to GATA-1, probes 2 and 3 containing individual CpG sites were incubated with nuclear extracts and compared with probe 1 for GATA-1 binding. (D) To determine the difference in binding affinity, binding of a radiolabeled unmethylated probe 1 was titrated with cold unmethylated and methylated probes.
Effect of methylated GATA elements on transcription activity in vitro
Next, we examined the effect of the CpG sites in GATA elements on transcription of CCR3 using a reporter assay (Figure 6A). The exon 1 sequence was cloned into a pGL3 basic vector and then subjected to in vitro methylation, which methylated all cytosines of CpG sites in the vector as well as in exon 1 (Hattori et al., 2004). When the methylated reporter plasmid was transfected into K562 cells, it completely lacked luciferase activity (data not shown), probably because methylation of CpG sites other than those in exon 1 completely nullified transcription of the reporter plasmid. To eliminate the effect resulting from methylation of the vector sequence itself, we directly cloned the exon 1 sequence with methylated or unmethylated CpG sites into the reporter plasmid. To do so, the exon 1 sequence was PCR-amplified, digested with two different restriction enzymes for the subsequent cloning procedure, and methylated in the absence or presence of S-adenosylmethionine. Both unmethylated and methylated exon 1 sequences were ligated to pGL3 basic vector. The whole ligation mixture was then transfected into K562 cells. When increasing concentrations of the exon 1 fragment of unmethylated CpG sites were ligated, transcription activity of the ligation mixture increased in proportion to the input amount of the exon 1 sequence used as an insert (Figure 6B). In addition, when the exon 1 fragment with a mutated version (mut1) of the first GATA site, which was shown to decrease transcription by more than one-half in K562 cells (Kim et al., 2010) and A549 cells (Supplemental Data Figure S1), was ligated to a pGL3 basic vector, its transcription activity was reduced by more than half compared to that of exon 1 with unmethylated CpG sites. These results ensure that the ligation mixture contains the proper recombinant plasmid for transcription activation. Exon 1 with methylated CpG sites in the GATA elements yielded a small but significant increase (125 ± 7.5%, P = 0.049) in transcription activity compared to that of unmethylated CpG sites. The transcription activity in the exon 1 was comparable to that of mut4-5, in which 5'-gatc-3' was replaced with 5'-tcga-3', presumably having an equivalent effect on exon 1 with methylated CpG in terms of inability to bind GATA-1 (Figure 6C). This result suggests that methylation of CpG sites in GATA elements in exon 1 can modulate the transcription activity of CCR3. We also created a point mutant in which the CG sequence within the CREB element was replaced with AT so that the methylation effect of the CpG site, if any, would be removed. The mutant had no effect on the transcription of the CCR3 reporter plasmid (data not shown).
Transcription activity of exon 1 with unmethylated and methylated CpG sites. (A) Constructs of exon 1 with methylated or unmethylated CpG sites at GATA elements. Methylated exon 1 was generated by in vitro methylation. Mut4-5, in which two TATC sequences were replaced with GCGA, which was expected to have an equivalent effect to those of methylated GATA elements, was used as a reference. Mut1, whose transcription activity was previously shown to be reduced by more than half compared with that of an unmutated version (Kim et al., 2010), was also used as a reference to validate this assay. Filled boxes represent GATA elements, and open and closed lollipops represent unmethylated and methylated CpG sites, respectively. U and M indicate unmethylated and methylated exon 1, respectively. (B) Varying concentrations of the unmethylated exon 1 fragment were ligated to a pGL3 basic vector. The whole ligation mixture was then transfected into K562 cells. Transfection efficiency was normalized by co-transfected renilla luciferase activity. The transcription activities were determined as relative values (%) compared to the transcription activity of the ligation mixture with the molar ratio of 5 unmethylated exon 1 (insert) to 1 vector. The data were expressed as mean ± SEM of two independent experiments performed in triplicate. (C) Methylated and unmethylated exon 1 were ligated with pGL3 basic vector (5:1 ratio), and their effects on transcription activity were determined. Transcription activity of methylated exon 1 was expressed relative to that of unmethylated exon 1, which was assumed to be 100%. The results represent the mean ± SEM of six independent experiments performed in triplicate.
Discussion
Exon 1, the major regulatory region for CCR3 transcription, includes a positively acting GATA element that is solely responsible for GATA-1-mediated transcription of the gene and two negatively acting GATA elements that are able to suppress CCR3 transcription in vitro. Since the negatively acting GATA elements contain CpG sites, we evaluated a correlation between methylation of these two sites and CCR3 mRNA level by bisulfate-pyrosequencing technology and real-time PCR with cells which express varying CCR3 mRNA levels, including peripheral blood eosinophils, CB-derived eosinophils, PBMC, and a panel of cells lines. Primary cells, including peripheral blood eosinophils, CB-derived eosinophils, and PBMC, expressed much higher level of CCR3 mRNA than PBMC and seven cell lines tested and had high methylation at the fourth and fifth GATA elements. Among cell lines, U937 and Jurkat cells, which constitutively express CCR3 mRNA, had high methylation statuses at these two GATA elements. Epithelial cell lines, especially A549 and 293T cells, expressed lower level of CCR3 mRNA than U937 and Jurkat cells, and had intermediate level of methylation at these two GATA elements. In contrast, two myeloid cell lines, K562 and HL-60 cells, which do not constitutively express CCR3 mRNA, exhibited little methylation (Table 1). Thus, DNA methylation of the two CpG sites at the GATA sites correlated with CCR3 mRNA expression in those cells. In line with the observations, the negatively acting GATA elements with methylated CpG sites had remarkably reduced binding to GATA-1 (Figure 5) and resulted in an increase in GATA-1-mediated transcription of CCR3 gene (Figure 6), possibly by redirecting available GATA-1 to the positive GATA element. These results show correlation of CCR3 mRNA expression with methylation of the CpG sites. Therefore, these results suggest that DNA methylation of these GATA elements may serve as an important epigenetic component of CCR3 transcription in vivo.
Despite the association of methylation of the CpG sites at GATA elements in the regulatory region of CCR3 with its transcription (Figure 4), the methylation effect on CCR3 transcription was significant but modest (Figure 6C). The reason for the modest increase appears to be largely attributed to the fact that the negatively acting GATA elements themselves do not influence CCR3 transcription as strongly as does the positively acting GATA element. In fact, a point mutant of the positively acting GATA element (Supplemental Data) and a mutant missing the GATA element exhibited severely impaired transcription (Kim et al., 2010), whereas a double mutant that disrupts both negatively acting GATA elements affected transcription in the reporter assay, but its effect was not as great as that of the positively acting GATA element (Figure 6C). In this context, methylation of CpG sites at the negatively acting GATA elements, while being thought to have an equivalent effect to the double mutant, is not likely to have a profound effect on CCR3 transcription in the cell, unlike the mutant lacking the positively acting GATA element.
GATA sequences and the complementary TATC sequences occur abundantly in the genome, GATA consensus sequences are found in the promoters and enhancers of many genes, and increasing numbers of GATA-1 target genes are being identified. They include genes that are indispensable for erythrocyte differentiation and functions, notably globin genes (Ferreira et al., 2005). DNA methylation ubiquitously occurs in globin gene clusters (Van der Ploeg and Flavell, 1980) and serves as a dominant mechanism to repress some globin genes (Goren et al., 2006). In addition, the regulatory regions of eosinophil-specific genes contain functional GATA elements (Dyer and Rosenberg, 2000; Du et al., 2002; Qiu et al., 2009), although methylation within these elements is not determined. Our results show considerably reduced binding affinity of GATA-1 for the methylated GATA element (Figure 5). These data suggest that if methylation of a CpG site within a GATA element exerts a crucial function for GATA-1-mediated transcription, methylation of the GATA element may greatly affect transcription of the gene. Many transcription factors are known to bind CpG-containing sequences, and most of them fail to bind and have reduced activity when the CpG site is methylated. These include CREB (Kim and Leonard, 2007), Myc (Prendergast et al., 1991; Perini et al., 2005), USF-1 (Aoki et al., 2008), AP-2 (Comb and Goodman, 1990), and CTCF (Bell and Felsenfeld, 2000). In contrast, other transcription factors, such as Sp1 or CAAT box-binding transcription factor (CTF), are able to bind to their binding elements regardless of methylation (Holler et al., 1988; Hantusch et al., 2007; Sunahori et al., 2009). Our results reveal that GATA-1 is a transcription factors sensitive to DNA methylation for binding and activity. Although such genes whose transcriptions are affected by DNA methylation at GATA elements are yet to be identified, the abundant occurrence of GATA consensus sequences in the genome raises the possibility that genes with GATA elements functioning as crucial cis-acting sequences may be transcriptionally regulated in a methylation-dependent manner.
One intriguing observation is that the in vitro differentiated eosinophils resemble peripheral blood eosinophils not only in a cellular phenotype (MBP expression level), but also in the methylation statuses of all the three CpG sites in exon 1 of CCR3. None of the cell lines, including eosinophilic cell line HL-60, were similar to terminally-differentiated CB-derived eosinophils in methylation pattern (Table 1). It is speculated that eosinophil progenitors with characteristic methylation statuses in these CpG sites might exist in an extremely small fraction within CD34+ cell population. The methylation pattern would actually remain unaltered throughout eosinophil development/differentiation, because the eosinophil progenitors selectively expand without changes in methylation and become a predominant population in the culture conditions favoring generation of eosinophils. Alternatively, the methylation pattern is progressively altered to become that of peripheral blood eosinophil at a single cell basis during eosinophil development/differentiation. It has recently been reported that modulation of CpG methylation occurs during lineage-specific differentiation in hematopietic cells. A notable instance, the myeloperoxidase gene, whose product serves as a marker for neutrophils, becomes progressively hypomethylated with upregulated expression during myeloid specification from multipotent progenitor to granulocyte/macrophage progenitor (Ji et al., 2010). Our results suggest that methylation of these CpG sites may serve as epigenetic marks during eosinophil development and differentiation, and it is likely to be one of the inherent elements accompanied with eosinophilopoiesis.
CB cells have provided a well-defined model to study eosinophil development and function. The parallel DNA methylation pattern of the CCR3 regulatory region between CB-derived eosinophils and peripheral blood eosinophils merits further investigation as to whether the coincidence extends to other well-known eosinophil-specific genes such as MBP, eosinophil peroxidase, eosinophil-derived neurotoxin, and eosinophil cationic protein. Analysis of CpGness shows that all of these genes have CpG-poor sequences at their proximal promoter regions, like the CCR3 gene. Furthermore, as gene microarray analysis of bone marrow-derived murine eosinophils failed to identify critical regulators that play a key role in eosinophil commitment and differentiation (Bystrom et al., 2004), analysis of a comprehensive methylation map of CB-derived eosinophils might help the understanding of epigenetic plasticity accompanying eosinophil restriction.
Methods
Ethics statement
Written informed consent was obtained from all subjects. The protocols used in this study were approved by the Soonchunhyang Bucheon Hospital's ethics committee (SCHBC-IRB-06-04).
Cell culture
A549 and HL-60 cells were purchased from Korean Cell Line Bank (Seoul, Korea), and K562, Jurkat, U937, NCI-H292, and 293T cells were obtained from ATCC (Manassas, VA). Human lung epithelial cells, A549 and NCI-H292, U937 cells, Jukat cells, HL-60 cells, and K562 cells were maintained in RPMI 1640 medium (Welgene, Seoul, Korea), and human embryo kidney 293T cells were maintained in DMEM (Welgene). All growth media were supplemented with 10% FBS, penicillin (100 U/ml), and streptomycin (100 µg/ml). Peripheral blood eosinophils were isolated from slightly atopic individuals. After RBC had been precipitated in 6% dextran-dextrose in 0.1 M EDTA (pH 7.4), the leukocyte-rich cell suspension was layered on a Percoll solution (1.070 g/ml) and centrifuged at 3,500 ×rpm for 30 min at 4℃. Enriched eosinophil fractions were incubated with anti-CD16 monoclonal antibody-conjugated microbeads (Miltenyi Biotec, Auburn, CA), and contaminating neutrophils were removed through negative selection using a MACS (BD PharMingen, San Diego, CA). Peripheral blood mononuclear cells were separated from blood obtained from healthy donors by density-gradient centrifugation over Ficoll-Paque Premium 1.073 (density, 1.077 g/ml, GE Healthcare, Uppsala, Sweden). CD34+ cells were immunomagnetically purified from human cord blood (CB) mononuclear cells using a MACS CD34+ microbead kit (Miltenyi Biotec). CD34+ cells were seeded in 24-well plates at 1×105 cells/well. The cells were cultured in IMDM (Welgene) containing 10% FBS, 100 U/ml penicillin, 100 µg/ml streptomycin supplemented with a cytokine cocktail of SCF (50 ng/ml), FLT-3L (50 ng/ml), GM-CSF (10 ng/ml), IL-3 (10 ng/ml), and IL-5 (10 ng/ml) for 6 d. Cells were then collected, split into 12-well plates at 1×106 cells/well in medium supplemented with IL-3 and IL-5, incubated with a half medium change for an additional 6 d, and incubated in medium supplemented with IL-5 for up to an additional 12 d with half medium changes every three day.
Flow cytometry
To determine MBP and CD34 expressions, cells were stained with an anti-human MBP (BD Pharmingen, San Diego, CA) or PE-conjugated anti-human CD34 antibody (BD Pharmingen). Isotype-matched antibody was also used to stain the cells. Cells stained with anti-human MBP were then incubated with a PE-conjugated anti-mouse IgG1 antibody (BD Pharmingen). Stained cells were analyzed by a FACSCaribur flow cytometer, and data analysis was performed using CellQuest Software (Becton Dickinson, San Jose, CA).
Bisulfite pyrosequencing
The exon 1 sequence of CCR3 was amplified using the forward primer and the biotinylated reverse primer designed by PSQ Assay Design (Biotage AB, Uppsala, Sweden). The primers for amplification were: for the CREB (+31) region, the forward primer was 5'-TGTTTGTGATTTGATGGTATTT-3' and the reverse primer was 5' (biotin)-AACTTCTATTATATTTCRATAAATTCTTACC-3'; for GATA (+103 and +118) elements, the forward primer was 5' (biotin)-GTAGGTATTATTGGTTTTTTTGTG-3' and the reverse primer was 5'-CCTCCTAAATCCAAAAACACT-3'. Twenty nanograms of genomic DNA was modified with sodium bisulfite using the EZ DNA Methylation kit (ZYMO Research, Irvine, CA) according to the manufacturer's instructions. Bisulfite-modified DNA was amplified in a 25 µl reaction with the primer set and 5 units of Taq polymerase (Solgent Co., Daejeon, Korea). The thermocycler program was as follows: 95℃ for 10 min; 40 cycles consisting of 95℃ for 45 s, 55℃ for 35 s, and 72℃ for 60 s; 72℃ for 10 min; and a hold at 4℃. For verification, the PCR products were visualized on a 1.5% agarose gel with ethidium bromide staining. Pyrosequencing reactions were carried out with sequencing primers on the PSQ HS 96A System (Biotage AB) according to the manufacturer's specifications. The methylation indexes of each gene promoter and of each sample were calculated as the average value of mC (mC+C) for all examined CpGs in the target region. Statistical correlations between MtI and the clinical variables recorded were calculated using SPSS. The primers for sequencing the CREB (+31) region were 5'-ATTTTTGTTTTAGGAGTG-3'; at GATA (+103 and +118) elements, 5'-TCTTCTAAATAAAAACTTCTATTATA-3'.
In vitro methylation and luciferase reporter assay
The exon 1 region of the CCR3 gene (-18/+179 relative to the transcription start site) was amplified using Primestar high fidelity Taq DNA polymerase (Takara, Shinga, Japan) and forward and reverse primers containing Xho I and Hind III restriction sites at the 5' ends, respectively. PCR primer sequences were previously described (Kim et al., 2010). PCR products were digested with Xho I and Hind III, and then the resulting Xho I-Hind III fragment was incubated with 24 units of M.SssI methylase (New England Biolabs, Ipswich, MA) in the presence (methylated) or absence (mock-methylated) of S-adenosylmethionine at 37℃ for 4 h. DNA methylation status was confirmed by digestion with methylation-sensitive Hpa II (New England Biolabs) (Kim et al., 2011). Methylated and mock-methylated DNA was ligated to pGL3-basic vector (Promega, Madison, WI). The total ligation mixture was purified with a PCR purification kit (Macherey-Nagel, Duren, Germany) and transfected with pRL-TK (Promega) vector using Lipofectamine 2000 (Invitrogen Life Technologies, Carlsbad, CA). After 36 h, luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega), and transfection efficiency was normalized to renilla luciferase activity.
Electrophoretic mobility shift assay (EMSA)
Nuclear extraction and EMSA were performed as previously described (Kim et al., 2010). Methylated and unmethylated oligonucleotides were synthesized (Bioneer Inc, Daejeon, Korea) and annealed with complementary oligonucleotides that were also methylated or unmethylated at cytosine residues within CpG sites. Nuclear extracts of K562 cells were incubated with 32P-labeled oligonucleotide probes in binding buffer (50 mM Tris-Cl [pH 7.5], 20% glycerol, 5 mM MgCl2, 2.5 mM EDTA, 2.5 mM DTT, 250 mM NaCl, and 0.25 mg/ml poly [dI-dC]·poly [dI-dC]). For supershift analysis, nuclear extracts were incubated with anti-GATA-1 (M-20, Santa Cruz Biotechnology, Santa Cruz, CA) or control antibody (Sigma-Aldrich, St. Louis, MO), followed by radiolabeled probes. The DNA-protein complexes were resolved on 6% nondenaturing polyacrylamide gels.
Immunofluorescence analysis
Cytospin was prepared from CD34+ cells and cultured with a cytokine cocktail for 24 days. Cells were fixed with 4% paraformaldehyde (Sigma-Aldrich) for 10 min, washed with PBS, and permeabilized with 0.1% saponin in PBS for 10 min. After washing three times with PBS, the cells were incubated in blocking buffer (3% BSA in PBS) for 1 h and then stained with FITC-conjugated anti-human MBP (BD Pharmingen) or FITC-conjugated anti-mouse IgG1 Ab (BD Pharmigen), and washed three times with PBS. All steps were carried out at room temperature. The cells were mounted in VECTASHIELD Mounting Medium with DAPI (Vector Laboratories, Burlingame, CA).
Real-time PCR
Total RNA was isolated using TRI reagent (Molecular Research Center, Cincinnati, OH) and treated with DNase I (Invitrogen Life Technologies). cDNA was reverse-transcribed from 2 µg of total RNA using SuperScript II RNase Reverse Transcriptase and random hexadeoxynucleotide primers (Invitrogen Life Technologies). Real-time PCR was performed with SYBR Green (Roche, Mannheim, Germany) mix using the ABI 7500 Real-Time PCR system (Applied Biosystems). Assays were carried out in triplicate. The primer sequences were as follows: CCR3 forward primer 5'-TCGTTCTCCCTCTGCTCG-3' and reverse primer 5'-CACCGCCATGATGACAAA-3'; GAPDH forward primer 5'-AGGGCTGCTTTTAACTCTGGT, and reverse primer 5'-CCCCACTTGATTTTGGAGGGA.
Statistical analysis
The data were managed and analyzed using SPSS 10.0 software (SPSS, Inc., Chicago, IL). To compare relative reporter activities between experiments, we used an independent t test. The correlation between the level of CCR3 mRNA expression and the degree of methylation on CpG was analyzed using Pearson correlation analysis. Statistical significance was defined at the standard 5% level with a two-tailed analysis.
Abbreviations
- CB:
-
cord blood
- MACS:
-
magnetic activated cell sorting
- MBP:
-
major basic protein
References
Aoki M, Terada T, Kajiwara M, Ogasawara K, Ikai I, Ogawa O, Katsura T, Inui K . Kidney-specific expression of human organic cation transporter 2 (OCT2/SLC22A2) is regulated by DNA methylation . Am J Physiol Renal Physiol 2008 ; 295 : F165 - F170
Beck LA, Tancowny B, Brummet ME, Asaki SY, Curry SL, Penno MB, Foster M, Bahl A, Stellato C . Functional analysis of the chemokine receptor CCR3 on airway epithelial cells . J Immunol 2006 ; 177 : 3344 - 3354
Bell AC, Felsenfeld G . Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene . Nature 2000 ; 405 : 482 - 485
Broske AM, Vockentanz L, Kharazi S, Huska MR, Mancini E, Scheller M, Kuhl C, Enns A, Prinz M, Jaenisch R, Nerlov C, Leutz A, Andrade-Navarro MA, Jacobsen SE, Rosenbauer F . DNA methylation protects hematopoietic stem cell multipotency from myeloerythroid restriction . Nat Genet 2009 ; 41 : 1207 - 1215
Bystrom J, Wynn TA, Domachowske JB, Rosenberg HF . Gene microarray analysis reveals interleukin-5-dependent transcriptional targets in mouse bone marrow . Blood 2004 ; 103 : 868 - 877
Comb M, Goodman HM . CpG methylation inhibits proenkephalin gene expression and binding of the transcription factor AP-2 . Nucleic Acids Res 1990 ; 18 : 3975 - 3982
Daugherty BL, Siciliano SJ, DeMartino JA, Malkowitz L, Sirotina A, Springer MS . Cloning, expression, and characterization of the human eosinophil eotaxin receptor . J Exp Med 1996 ; 183 : 2349 - 2354
Du J, Stankiewicz MJ, Liu Y, Xi Q, Schmitz JE, Lekstrom-Himes JA, Ackerman SJ . Novel combinatorial interactions of GATA-1, PU.1, and C/EBPepsilon isoforms regulate transcription of the gene encoding eosinophil granule major basic protein . J Biol Chem 2002 ; 277 : 43481 - 43494
Dyer KD, Rosenberg HF . Shared features of transcription: mutational analysis of the eosinophil/basophil Charcot-Leyden crystal protein gene promoter . J Leukoc Biol 2000 ; 67 : 691 - 698
Ferreira R, Ohneda K, Yamamoto M, Philipsen S . GATA1 function, a paradigm for transcription factors in hematopoiesis . Mol Cell Biol 2005 ; 25 : 1215 - 1227
Flood-Page PT, Menzies-Gow AN, Kay AB, Robinson DS . Eosinophil's role remains uncertain as anti-interleukin-5 only partially depletes numbers in asthmatic airway . Am J Respir Crit Care Med 2003 ; 167 : 199 - 204
Forsythe P, Befus AD . CCR3: a key to mast cell phenotypic and functional diversity ? Am J Respir Cell Mol Biol 2003 ; 28 : 405 - 409
Goren A, Simchen G, Fibach E, Szabo PE, Tanimoto K, Chakalova L, Pfeifer GP, Fraser PJ, Engel JD, Cedar H . Fine tuning of globin gene expression by DNA methylation . PLoS One 2006 ; 1 : e46
Haldar P, Brightling CE, Hargadon B, Gupta S, Monteiro W, Sousa A, Marshall RP, Bradding P, Green RH, Wardlaw AJ, Pavord ID . Mepolizumab and exacerbations of refractory eosinophilic asthma . N Engl J Med 2009 ; 360 : 973 - 984
Hattori N, Nishino K, Ko YG, Ohgane J, Tanaka S, Shiota K . Epigenetic control of mouse Oct-4 gene expression in embryonic stem cells and trophoblast stem cells . J Biol Chem 2004 ; 279 : 17063 - 17069
Hantusch B, Kalt R, Krieger S, Puri C, Kerjaschki D . Sp1/Sp3 and DNA-methylation contribute to basal transcriptional activation of human podoplanin in MG63 versus Saos-2 osteoblastic cells . BMC Mol Biol 2007 ; 8 : 20 -
Hirasawa R, Shimizu R, Takahashi S, Osawa M, Takayanagi S, Kato Y, Onodera M, Minegishi N, Yamamoto M, Fukao K, Taniguchi H, Nakauchi H, Iwama A . Essential and instructive roles of GATA factors in eosinophil development . J Exp Med 2002 ; 195 : 1379 - 1386
Holler M, Westin G, Jiricny J, Schaffner W . Sp1 transcription factor binds DNA and activates transcription even when the binding site is CpG methylated . Genes Dev 1988 ; 2 : 1127 - 1135
Ishihara K, Takahashi A, Kaneko M, Sugeno H, Hirasawa N, Hong J, Zee O, Ohuchi K . Differentiation of eosinophilic leukemia EoL-1 cells into eosinophils induced by histone deacetylase inhibitors . Life Sci 2007 ; 80 : 1213 - 1220
Iwama A, Osawa M, Hirasawa R, Uchiyama N, Kaneko S, Onodera M, Shibuya K, Shibuya A, Vinson C, Tenen DG, Nakauchi H . Reciprocal roles for CCAAT/enhancer binding protein (C/EBP) and PU.1 transcription factors in Langerhans cell commitment . J Exp Med 2002 ; 195 : 547 - 558
Ji H, Ehrlich LI, Seita J, Murakami P, Doi A, Lindau P, Lee H, Aryee MJ, Irizarry RA, Kim K, Rossi DJ, Inlay MA, Serwold T, Karsunky H, Ho L, Daley GQ, Weissman IL, Feinberg AP . Comprehensive methylome map of lineage commitment from haematopoietic progenitors . Nature 2010 ; 467 : 338 - 342
Kang JH, Lee DH, Lee JS, Kim HJ, Shin JW, Lee YH, Lee YS, Park CS, Chung IY . Eosinophilic differentiation is promoted by blockage of Notch signaling with a gamma-secretase inhibitor . Eur J Immunol 2005 ; 35 : 2982 - 2990
Kim BS, Uhm TG, Lee SK, Lee SH, Kang JH, Park CS, Chung IY . The crucial role of GATA-1 in CCR3 gene transcription: modulated balance by multiple GATA elements in the CCR3 regulatory region . J Immunol 2010 ; 185 : 6866 - 6875
Kim do N, Song YJ, Lee SK . The role of promoter methylation in Epstein-Barr virus (EBV) microRNA expression in EBV-infected B cell line . Exp Mol Med 2011 ; 43 : 401 - 410
Kim HP, Leonard WJ . CREB/ATF-dependent T cell receptor-induced FoxP3 gene expression: a role for DNA methylation . J Exp Med 2007 ; 204 : 1543 - 1551
Kitaura M, Nakajima T, Imai T, Harada S, Combadiere C . Tiffany HL, Murphy PM, Yoshie O. Molecular cloning of human eotaxin, an eosinophil-selective CC chemokine, and identification of a specific eosinophil eotaxin receptor, CC chemokine receptor 3 . J Biol Chem 1996 ; 271 : 7725 - 7730
Klose RJ, Bird AP . Genomic DNA methylation: the mark and its mediators . Trends Biochem Sci 2006 ; 31 : 89 - 97
Lamkhioued B, Abdelilah SG, Hamid Q, Mansour N, Delespesse G, Renzi PM . The CCR3 receptor is involved in eosinophil differentiation and is up-regulated by Th2 cytokines in CD34+ progenitor cells . J Immunol 2003 ; 170 : 537 - 547
Leckie MJ, Ten Brinke A, Khan J, Diamant Z, O'Connor BJ, Walls CM, Mathur AK, Cowley HC, Chung KF, Djukanovic R, Hansel TT, Holgate ST, Sterk PJ, Barnes PJ . Effects of an interleukin-5 blocking monoclonal antibody on eosinophils, airway hyper-responsiveness, and the late asthmatic response . Lancet 2000 ; 356 : 2144 - 2148
Li E . Chromatin modification and epigenetic reprogramming in mammalian development . Nat Rev Genet 2002 ; 3 : 662 - 673
McNagny K, Graf T . Making eosinophils through subtle shifts in transcription factor expression . J Exp Med 2002 ; 195 : F43 - F47
Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, Gnirke A, Jaenisch R, Lander ES . Genome-scale DNA methylation maps of pluripotent and differentiated cells . Nature 2008 ; 454 : 766 - 770
Nair P, Pizzichini MM, Kjarsgaard M, Inman MD, Efthimiadis A, Pizzichini E, Hargreave FE, O'Byrne PM . Mepolizumab for prednisone-dependent asthma with sputum eosinophilia . N Engl J Med 2009 ; 360 : 985 - 993
Nerlov C, Graf T . PU.1 induces myeloid lineage commitment in multipotent hematopoietic progenitors . Genes Dev 1998 ; 12 : 2403 - 2412
Okano M, Bell DW, Haber DA, Li E . DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development . Cell 1999 ; 99 : 247 - 257
Perini G, Diolaiti D, Porro A, Della Valle G . In vivo transcriptional regulation of N-Myc target genes is controlled by E-box methylation . Proc Natl Acad Sci USA 2005 ; 102 : 12117 - 12122
Ponath PD, Qin S, Post TW, Wang J, Wu L, Gerard NP, Newman W, Gerard C, Mackay CR . Molecular cloning and characterization of a human eotaxin receptor expressed selectively on eosinophils . J Exp Med 1996 ; 183 : 2437 - 2448
Prendergast GC, Lawe D, Ziff EB . Association of Myn, the murine homolog of max, with c-Myc stimulates methylation-sensitive DNA binding and ras cotransformation . Cell 1991 ; 65 : 395 - 407
Qiu Z, Dyer KD, Xie Z, Radinger M, Rosenberg HF . GATA transcription factors regulate the expression of the human eosinophil-derived neurotoxin (RNase 2) gene . J Biol Chem 2009 ; 284 : 13099 - 13109
Rådinger M, Lötvall J . Eosinophil progenitors in allergy and asthma do they matter ? Pharmacol Ther 2009 ; 121 : 174 - 184
Reik W . Stability and flexibility of epigenetic gene regulation in mammalian development . Nature 2007 ; 447 : 425 - 432
Rothenberg ME, Hogan SP . The eosinophil . Annu Rev Immunol 2006 ; 24 : 147 - 174
Sallusto F, Mackay CR, Lanzavecchia A . Selective expression of the eotaxin receptor CCR3 by human T helper 2 cells . Science 1997 ; 277 : 2005 - 2007
Scotet E, Schroeder S, Lanzavecchia A . Molecular regulation of CC-chemokine receptor 3 expression in human T helper 2 cells . Blood 2001 ; 98 : 2568 - 2570
Stellato C, Brummet ME, Plitt JR, Shahabuddin S, Baroody FM, Liu MC, Ponath PD, Beck LA . Expression of the C-C chemokine receptor CCR3 in human airway epithelial cells . J Immunol 2001 ; 166 : 1457 - 1461
Sunahori K, Juang YT, Tsokos GC . Methylation status of CpG islands flanking a cAMP response element motif on the protein phosphatase 2Ac alpha promoter determines CREB binding and activity . J Immunol 2009 ; 182 : 1500 - 1508
Tiffany HL, Alkhatib G, Combadiere C, Berger EA, Murphy PM . CC chemokine receptors 1 and 3 are differentially regulated by IL-5 during maturation of eosinophilic HL-60 cells . J Immunol 1998 ; 160 : 1385 - 1392
Trowbridge JJ, Snow JW, Kim J, Orkin SH . DNA methyltransferase 1 is essential for and uniquely regulates hematopoietic stem and progenitor cells . Cell Stem Cell 2009 ; 5 : 442 - 449
Van der Ploeg LH, Flavell RA . DNA methylation in the human gamma delta beta-globin locus in erythroid and nonerythroid tissues . Cell 1980 ; 19 : 947 - 958
Vijh S, Dayhoff DE, Wang CE, Imam Z, Ehrenberg PK, Michael NL . Transcription regulation of human chemokine receptor CCR3: evidence for a rare TATA-less promoter structure conserved between drosophila and humans . Genomics 2002 ; 80 : 86 - 95
Willems LI, Ijzerman AP . Small molecule antagonists for chemokine CCR3 receptors . Med Res Rev 2010 ; 30 : 778 - 817
Yu C, Cantor AB, Yang H, Browne C, Wells RA, Fujiwara Y, Orkin SH . Targeted deletion of a high-affinity GATA-binding site in the GATA-1 promoter leads to selective loss of the eosinophil lineage in vivo . J Exp Med 2002 ; 195 : 1387 - 1395
Zhang DE, Zhang P, Wang ND, Hetherington CJ, Darlington GJ, Tenen DG . Absence of granulocyte colony-stimulating factor signaling and neutrophil development in CCAAT enhancer binding protein alpha-deficient mice . Proc Natl Acad Sci USA 1997 ; 94 : 569 - 574
Zhang Y, Rohde C, Tierling S, Jurkowski TP, Bock C, Santacruz D, Ragozin S, Reinhardt R, Groth M, Walter J, Jeltsch A . DNA methylation analysis of chromosome 21 gene promoters at single base pair and single allele resolution . PLoS Genet 2009 ; 5 : e1000438
Zhao Z, Han L . CpG islands: algorithms and applications in methylation studies . Biochem Biophys Res Commun 2009 ; 382 : 643 - 645
Zimmermann N, Daugherty BL, Kavanaugh JL, El-Awar FY, Moulton EA, Rothenberg ME . Analysis of the CC chemokine receptor 3 gene reveals a complex 5' exon organization, a functional role for untranslated exon 1, and a broadly active promoter with eosinophil-selective elements . Blood 2000 ; 96 : 2346 - 2354
Zimmermann N, Colyer JL, Koch LE, Rothenberg ME . Analysis of the CCR3 promoter reveals a regulatory region in exon 1 that binds GATA-1 . BMC Immunol 2005 ; 6 : 7
Acknowledgements
Authors greatly appreciate Dr. Sung Hee Choi (Hanvik Clinic, Ansan, Korea) for providing experimental materials. This work was supported by a grant 2011-0014580 (to IYC) from the National Research Foundation, Republic of Korea. BSK and JHK were supported by BK21, National Research Foundation. The authors have no financial conflicts of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on the Experimental & Molecular Medicine website
Supplementary information
Rights and permissions
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Uhm, T., Lee, S., Kim, B. et al. CpG methylation at GATA elements in the regulatory region of CCR3 positively correlates with CCR3 transcription. Exp Mol Med 44, 268–280 (2012). https://doi.org/10.3858/emm.2012.44.4.022
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.3858/emm.2012.44.4.022
Keywords
This article is cited by
-
Aspirin induces IL-4 production: augmented IL-4 production in aspirin-exacerbated respiratory disease
Experimental & Molecular Medicine (2016)