Abstract
It is not yet understood how the enhanced expression of pancreatic adenocarcinoma up-regulated factor (PAUF; a novel oncogene identified in our recent studies), contributes to the oncogenesis of pancreatic cells. We herein report that PAUF up-regulates the expression and transcriptional activity of β-catenin while the suppression of PAUF by shRNA down-regulates β-catenin. The induction of β-catenin by PAUF is mediated by the activities of Akt and GSK-3β, but inhibition of downstream ERK does not reduce β-catenin expression. To test whether PAUF emulates either the Wnt3a-mediated or the protein kinase A-mediated signaling pathway for the stabilization of β-catenin, we examined the phosphorylation status of β-catenin in the presence of PAUF compared with that of β-catenin during treatment with Wnt3a or dibutyryl cAMP, a cell permeable cyclic AMP analogue. PAUF expression induces phosphorylation at Ser-33/37/Thr-41 and Ser-675 of β-catenin but no phosphorylation at Ser-45, indicating that a unique phosphorylation pattern of β-catenin is caused by PAUF. Finally, the expression of PAUF up-regulates both cyclin-D1 and c-Jun, target genes of β-catenin, leading to a rapid proliferation of pancreatic cells; conversely decreased PAUF expression (by shRNA) results in the reduced proliferation of pancreatic cells. Treatment with hexachlorophene (an inhibitor of β-catenin) reduces the proliferation of pancreatic cells despite the presence of PAUF. Taken together, we propose that PAUF can up-regulate and stabilize β-catenin via a novel pattern of phosphorylation, thereby contributing to the rapid proliferation of pancreatic cancer cells.
Similar content being viewed by others
Introduction
Pancreatic cancer has an extremely poor prognosis with a 5-yr survival rate of less than 5% (Ghaneh et al., 2007; Maitra and Hruban, 2008). The only potential curative treatment for pancreatic cancer is surgery, but only 10-20% of patients are candidates for surgery at the time of presentation. To overcome obstacles such as late diagnosis, the aggressive nature of the tumor, and resistance to existing therapeutic regimens (Hawes et al., 2000), novel tumor-specific biomarkers and genetic pathways that may lead to potential diagnostic and therapeutic targets are desperately needed.
Studies of gene expression patterns using serial analysis of gene expression (SAGE), cDNA arrays and oligonucleotide arrays provide much information about the molecular characteristics of pancreatic cancers and can be used to distinguish closely related cancer subtypes (Iacobuzio-Donahue et al., 2003a; 2003b). In a recent study, we reported that a novel secretory protein, pancreatic adenocarcinoma up-regulated factor (PAUF), is highly expressed not only in pancreatic cancer tissues but also in colon, ovary and stomach cancer tissues and the introduction of PAUF into CHO cells causes tumor formation in xenografted nude mice (Kim et al., 2009). In further studies, we demonstrated that PAUF expression enhances migration and invasion by up-regulating CXCR4, which eventually facilitates the metastasis of pancreatic cancer (Lee et al., 2010).
Wnts are secreted glycoproteins that play important roles in cell proliferation, differentiation and oncogenesis (Polakis, 2000). The Wnt signal is initiated by the interaction of Wnts (Wnt1, 3a, and 8) with the Frizzled receptor and low density lipoprotein receptor-related protein 5/6 (LRP 5/6) co-receptors (Giles et al., 2003). The signal is then transduced through the disheveled protein to negatively regulate glycogen synthase kinase-3β (GSK-3β), resulting in the accumulation of cytoplasmic β-catenin. The β-catenin is then translocated into the nucleus, where it forms a complex with the T cell factor/lymphocyte enhancer factor (TCF/LEF) families of transcription factors to activate the expression of β-catenin responsive genes, such as cyclin-D1, c-Jun, c-Myc and peroxisome proliferator-activated receptor-δ (He et al., 1998, 1999; Tetsu and McCormick, 1999; Saadeddin et al., 2009). Many types of cancers display the accumulation of β-catenin and the consequent activation of TCF/LEF-dependent gene transcription.
In quiescent cells, β-catenin is maintained in the cytoplasm at low levels. This is facilitated by its interaction with scaffolding proteins, adenomatous polyposis coli (APC) protein, axin, and protein kinases such as casein kinase 1a (CK1a) and GSK-3β, which phosphorylate β-catenin on Ser-45 and then Ser-33/37/Thr-41 sites, respectively, leading to its ubiquitination and proteosomal degradation (Rubinfeld et al., 1996; Aberle et al., 1997; Liu et al., 2002). Wnt and other growth stimuli cause the phosphorylation of GSK-3β, resulting in the inactivation of GSK-3β-mediated phosphorylation at the Ser-33/37/Thr-41 sites and then culminating in the stabilization of β-catenin and subsequent translocation to the nucleus (van Noort et al., 2002). Other recent studies have shown that Protein Kinase A (PKA) can also stabilize β-catenin, in this case by phosphorylation at the Ser-675 site (Hino et al., 2005; Taurin et al., 2006).
Following our earlier demonstration that PAUF can contribute to the oncogenesis of pancreatic cells, we herein propose a mechanism for this effect, which is that PAUF can induce the up-regulation of β-catenin and β-catenin-mediated transcriptional activity, which may in turn potentiate malignant progression in pancreatic cells.
Results
PAUF up-regulates the expression and transcriptional activity of β-catenin
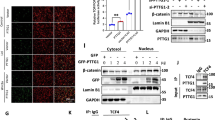
In earlier work we described an up-regulated novel gene in pancreatic cancer cells (which we called pancreatic adenocarcinoma up-regulated factor; PAUF) and showed its oncogenic activity using a xenografted nude mouse model (Kim et al., 2009). However, the mechanism by which PAUF could contribute to the oncogenesis of pancreatic cells was not clear. Our attention was drawn to β-catenin, which plays a crucial role in many biological phenomena such as development, cell proliferation and tumorigenesis (Ghaneh et al., 2007; Maitra and Hruban, 2008). To explore whether PAUF might regulate β-catenin, we first transfected the Panc-1 pancreatic cell line so that it stably contained either a vector alone (Panc-Vec) or vector plus PAUF (Panc-PAUF), and BxPC3, pancreatic cells stably expressing either a control shRNA (Bx-ConshRNA) or PAUF shRNA (Bx-PAUFshRNA). As shown in Figure 1A, Panc-PAUF cells dramatically enhanced the expression of β-catenin compared with Panc-Vec. In contrast, Bx-PAUFshRNA cells showed reduced β-catenin levels compared with Bx-ConshRNA cells. When we furthermore examined the level of β-catenin mRNA in Panc-PAUF and Bx-PAUFshRNA cells, we found that the stable expression of PAUF also elevated β-catenin transcripts in Panc-PAUF cells compared with those in Panc-Vec cells while reduction of PAUF diminished β-catenin transcripts in Bx-PAUFshRNA cells compared with those in Bx-ConshRNA cells (Figure 1B). In addition, we introduced a Top Flash luciferase reporter vector, controlled by β-catenin/TCF-4 complex, to examine whether the elevated β-catenin levels caused by PAUF lead to functional activation and binding to the β-catenin/TCF binding element in the nucleus. As seen in Figure 1C, PAUF transfection enhanced β-catenin-mediated transcriptional activity in HEK293 T cells in a dose-dependent manner while PAUF transfection together with Fop Flash vector carrying a mutant TCF binding site failed to enhance β-catenin-mediated transcription activity. Taken together, these results suggest that PAUF expression induces not only β-catenin transcriptional activity but also up-regulation of the β-catenin protein level and activation of its downstream targets.
PAUF induces the enhancement of β-catenin protein and transcripts, leading to up-regulation of its transcriptional activity. (A) Panc-Vec (Con), Panc-PAUF (PAUF), Bx-ConshRNA (Con-Sh), and Bx-PAUFshRNA (PAUF-Sh) cells were prepared for cell lysates. Proteins were separated by 10% SDS-polyacrylamide gel electrophoresis (PAGE) and the expression of β-catenin was examined by immunoblotting using anti-β-catenin antibody. (B) The cells were harvested and total RNA was isolated followed by reverse transcription. β-catenin sequences were amplified with specific primers and visualized on 1.5% agarose gels after ethidium bromide staining. β-actin was used for an internal control. (C) HEK293 T cells were transfected with Top-Flash (0.2, 1 µg) or Fop-Flash (0.2, 1 µg) luciferase reporter vector and were harvested at 48 h post-transfection. When the luciferase activity was measured, transfection efficiency was normalized with a β-galactosidase reporter vector pGK-β-gal (1 µg). The results shown are the average of three experiments; bars indicate standard deviations.
PAUF activates Akt-GSK3β signaling via a unique pattern of phosphorylation of β-catenin
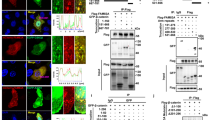
To determine whether PAUF-mediated up-regulation of β-catenin emulates activation patterns induced through the Wnt3a signaling pathway, we employed HEK293 cells expressing Frizzled-1 (HEK293-FZ), a Wnt3a receptor. The cells were treated with Wnt3a, and we compared the phosphorylation pattern of Akt and GSK-3β with that of Akt and GSK-3β in HEK293-Fz cells that were instead transfected with a PAUF vector. We also included in this analysis HEK293-Fz cells treated with Bt2-cAMP, as it has been reported that Protein kinase A (PKA) activation using this cAMP analogue induces the accumulation of β-catenin (Hino et al., 2005). As seen in Figure 2A, both Wnt3a and Bt2-cAMP treatments elevated phosphorylation of Akt and especially GSK-3β; this was also seen in the PAUF-transfected cells, although the degree of activation of GSK-3β is much less. To dissect the extent to which PAUF-mediated signaling really overlaps with Wnt3a- or Bt2-cAMP-mediated signaling, we compared the detailed phosphorylation status of β-catenin after PAUF transfection with that of β-catenin during Wnt3a or Bt2-cAMP treatment. Previous studies have shown that the canonical regulation of β-catenin is achieved through its phosphorylation by CK1α at the Ser-45 site and by GSK-3β at the Sre-33/37 and Thr-41 sites. These phosphorylation events target β-catenin for ubiquitination and subsequent degradation through the proteosome system (Rubinfeld et al., 1996; Aberle et al., 1997; Liu et al., 2002). In contrast, recent studies have reported that PKA can phosphorylate β-catenin at Ser-552 and Ser-675, resulting in its stabilization (Hino et al., 2005; Taurin et al., 2006). Thus, we wondered whether PAUF expression would modulate phosphorylation of β-catenin through the canonical Wnt3a signaling pathway or the noncanonical PKA pathway. As shown in Figure 2B, all three treatments (Wnt3a, Bt2-cAMP and PAUF transfection) caused a significant accumulation of β-catenin protein; however Wnt3a and Bt2-cAMP treatment actually decreased phosphorylation of the Ser- 33/37 and Thr-41 residues while PAUF expression by transfection slightly increased phosphorylation at these residues. In addition, PAUF expression greatly decreased phosphorylation of Ser-45 while Wnt3a signaling and Bt2-cAMP treatment caused a minor decrease in phosphorylation at the Ser-45. When we examined the extent of phosphorylation at the Ser-675 residue during each treatment, we found that all three stimuli resulted in an increased phosphorylation signal that corresponded to the increase of β-catenin levels contrary to earlier reports (Hino et al., 2005; Taurin et al., 2006). Taken together, although each of PAUF, Wnt3a and Bt2-cAMP utilizes the Akt-GSK3β pathway, it seems that PAUF may activate a distinct phosphorylation pattern for β-catenin regulation.
PAUF activates the Akt-GSK-3β signaling axis but exhibits a different phosphorylation pattern of β-catenin compared with Wnt3a- or Bt2-cAMP treatment. (A, B) HEK293-FZ cells were treated with culture medium containing Wnt3a or culture medium without Wnt3a as a vehicle, and Bt2-cAMP or DMSO as a vehicle. HEK293-Fz cells were also transfected with PAUF expression vector or an empty vector. After proteins from cell lysates were separated on 10% SDS-PAGE, Akt, GSK-3β, β-catenin and PAUF were detected with the corresponding antibodies and their activities were measured with the corresponding phospho-specific antibodies. Phosphoryation states of β-catenin after the treatment were detected with anti-phospho-β-catenin antibodies for Ser-33/37 and Thr-41, Ser-45, and Ser-675 of β-catenin. Band intensities of phospho-β-catenin were analyzed with Multi Gauge Ver2.1 program. Relative intensity of the sample was expressed compared with that of mock-treated control in each group.
PAUF utilizes the Akt-GSK-3β pathway but does not use PKA, Ras or ERK MAPK signaling molecules for the up-regulation of β-catenin
We next wondered how PAUF is able to up-regulate and activate β-catenin. In particular, we questioned whether the Akt-GSK-3β or the PKA signaling pathways are specifically involved in the regulation of β-catenin expression and activity. To answer this question, we treated Panc-Vec or Panc-PAUF cells with wortmannin (an inhibitor of PI-3 kinase and indirectly of Akt), LiCl (an inhibitor of GSK-3β) or H-89 (an inhibitor of PKA). As shown in Figure 3A, PI-3 kinase/Akt inhibition by wortmannin reduced β-catenin protein levels while GSK-3β inhibition by LiCl enhanced the abundance of β-catenin protein, even after PAUF stimulation. H-89 treatment did not affect the β-catenin protein level in Panc-PAUF cells. These results at least suggest that PAUF utilizes the Akt-GSK-3β signaling pathway for the regulation of β-catenin. As earlier studies have reported that activation of ERK and Ras positively regulate Wnt3a signaling and lead to β-catenin stabilization (Kim and Choi, 2007; Castellone et al., 2009), we next examined Ras activity in Panc-PAUF cells. As seen in Figure 3B, similar levels of active Ras molecules are present in both Panc-Vec and Panc-PAUF cells as detected by binding to GST-Raf-RBD fusion protein, indicating that PAUF does not activate Ras. As our recent study has shown that MAPKs (including ERK and JNK) are activated during soluble PAUF protein treatment (Lee et al., 2010), we then treated Panc-Vec or Panc-PAUF cells with MAPK inhibitors to assess a possible role for MAPK in PAUF-mediated β-catenin up-regulation. Of interest, we found that ERK inhibition does not influence β-catenin expression in the presence or absence of PAUF, while inhibition of JNK or p38 MAPK mildly represses PAUF-induced β-catenin expression (Figure 3C), suggesting that PAUF-mediated signaling for β-catenin up-regulation is different from that of Wnt3a.
Akt-GSK-3β signaling is involved in PAUF-mediated stabilization of β-catenin but PKA, Ras, and ERK are not. (A) Panc-Vec and Panc-PAUF cells were treated with wortmannin (10 µM), LiCl (20 mM), or H-89 (1 mM) and DMSO as mock for 12 h and then prepared as cell lysates. After proteins from cell lysates were separated on 10% SDS-PAGE, β-catenin and β-tubulin were detected with the corresponding antibodies. (B) Glutathione S-transferase (GST) or GST-Raf-RBD protein were added to cell lysates from Panc-Vec and Panc-PAUF and active Ras was pulled down with glutathione-agarose. The precipitated mixtures were separated on 12% SDS-PAGE and Ras protein was detected with anti-Ras antibody. Total Ras protein in the lysates was also detected (bottom center) in the lysates as a control. (C) Panc-Vec and Panc-PAUF cells were treated with PD98059 (PD; 30 µM), SP600125 (SP; 20 µM), or SB202190 (SB; 20 µM) MAPK inhibitors and harvested at 12 h after treatment. After proteins from cell lysates were separated on 10% SDS-PAGE, β-catenin, ERK, JNK, and p38 MAPK were detected with the corresponding antibodies and their activities were measured with the corresponding phospho-specific antibodies.
PAUF enhances the expression of cyclin D1 and c-Jun, target genes of β-catenin, resulting in a rapid proliferation of pancreatic cells
To explore possible biological consequences of β-catenin up-regulation as caused by PAUF, we examined the expression levels of cyclin D1 and c-Jun, which are known as target genes of β-catenin. As shown in Figures 4A and 4B, the stable expression of PAUF increased cyclin D1 protein levels in Panc-PAUF cells compared with those in Panc-Vec cells, while the shRNA-mediated reduction of PAUF diminished c-Jun protein levels in Bx-PAUFshRNA cells compared with that in Bx-ConshRNA cells. Furthermore, when we counted cell numbers over three days in Panc-PAUF and Panc-Vec cells to determine the consequences of up-regulation of cyclin D1 and c-Jun mediated by PAUF, we observed that Panc-PAUF cells proliferate slightly more rapidly than Panc-Vec cells (P < 0.05, Figure 4A). Conversely, when we counted cell numbers in Bx-ConshRNA and Bx-PAUFshRNA cells, we clearly showed that Bx-PAUFshRNA cells exhibited slower proliferation than Bx-ConshRNA cells (P < 0.01, Figure 4B). These results suggest that up-regulation of cyclin D1 and c-Jun mediated by PAUF can accelerate pancreatic cell proliferation. Next, to address the possibility that the expression levels of β-catenin are directly related to the proliferation of pancreatic cells, hexachlorophene (an inhibitor of β-catenin that was used for our previous study; Min et al., 2009), was used to treat Panc-PAUF and Bx-ConshRNA cells. As seen in Figure 4C, inhibition of β-catenin expression with hexachlorophene suppressed both cell lines despite the presence of PAUF (P < 0.01). This result indicates that PAUF expression most likely enhances pancreatic cell proliferation through β-catenin.
Up-regulation of β-catenin mediated by PAUF accelerates proliferation of pancreatic cells. (A) Panc-Vec (Con) and Panc-PAUF (PAUF) cells were seeded on 12 well plates at 1 × 105 cells per well and cultured for 72 h. The cell numbers were counted at 24, 48 and 72 h using Trypan blue staining. Panc-Vec and Panc-PAUF cells were prepared for cell lysates. Proteins were separated from the cell lysates on 10% SDS-PAGE and the expression of β-catenin and cyclinD1 were examined by immunoblotting using the corresponding antibodies. The results shown are the average of three experiments; bars indicate standard deviations. (*, P < 0.05) (B) Proliferation of Bx-ConshRNA (Con-Sh) and Bx-PAUFshRNA (PAUF-Sh) cells and the expression of β-catenin and c-Jun were analyzed as described in Figure 4A. The results shown are the average of three experiments; bars indicate standard deviations. (**, P < 0.01). (C) Panc-PAUF and Bx-ConshRNA cells were seeded on 12 well plates at 1 × 105 cells per well and then treated with hexachlorophene (10 µM) for 48 h. The cell numbers was counted at 48 h using Trypan blue staining. The results shown are the average of triplicate wells. Bars indicate standard deviations (**, P < 0.01). The expression of β-catenin was detected as described in Figure 4A.
Discussion
Pancreatic cancer has a high mortality rate and short survival, as a result of the high incidence of metastatic disease at diagnosis, the fulminant clinical course and the lack of successful therapeutic strategies (Hawes et al., 2000). We therefore sought in an earlier study to find useful diagnostic biomarkers or therapeutic molecular targets using a genechip expression analysis (Kim et al., 2009). This study led us to discover a novel gene that we named pancreatic adenocarcinoma up-regulated factor (PAUF), which was highly expressed in human pancreatic cancer. Further study revealed that PAUF plays important roles in cancer progression such as oncogenic activity and metastasis (Kim et al., 2009; Lee et al., 2010), and we have argued that PAUF may be a novel diagnostic marker as well as a therapeutic target in pancreatic cancer.
However, the molecular mechanism by which PAUF participates in normal cell signaling and when in excess mediates the development of pancreatic cancer both remain unknown. We herein report that PAUF specifically activates and stabilizes β-catenin, leading to the rapid proliferation of pancreatic cell lines. We find that PAUF-induced phosphorylation of β-catenin follows a different phosphorylation pattern compared with that achieved by Wnt3a or Bt2-cAMP treatment. Surprisingly, despite the common usage of Akt-GSK3β signaling by PAUF, Wnt3a or Bt2-cAMP, only PAUF induced β-catenin hyper-phosphorylation of Ser-33/37 and Thr-41, which is thought to lead to its ubiquination and proteosomal degradation, but in this case β-catenin was actually stabilized. However, Wnt3a and Bt2-cAMP did not induce β-catenin phosphorylation at the Ser-33/37 and Thr-41 residues. On the basis of these results, we speculate that PAUF may also recruit or activate a molecule that is downstream of GSK-3β, and thereby interrupt the ubiquitin E3 ligase-mediated degradation of β-catenin that is otherwise predicted. For example, there is growing evidence that CK2 phosphorylates Thr-393 of β-catenin, leading to its stabilization (Wu et al., 2009). In addition, a long form of cellular FLIP (FLICE/caspase 8 inhibitory protein) has been shown to stabilize β-catenin (Nakagiri et al., 2005). Furthermore, it has been recently reported that the acetyltransferase p300/CBP-associated factor (PCAF) can directly acteylate and stablize β-catenin, leading to its stabilization and, finally, protease-activated receptor-1 (PAR-1) can also induce β-catenin stabilization through Gα13, independently of Wnt signaling (Ge et al., 2009). Thus, in our next round of experiments, we will explore in more detail the molecular mechanisms by which PAUF signaling can stabilize and activate β-catenin.
Our observation that β-catenin stabilization can enhance the proliferation of pancreatic cell lines is strongly supported by other studies showing that activation of β-catenin alone is sufficient to induce pancreatic tumorigenesis using Cre/lox transgenic mice (Heiser et al., 2008), and also that deletion of the β-catenin gene results in a loss of pancreas mass during development (Heiser et al., 2006). We suggest that the rapid proliferation of pancreatic cancer cell lines can be explained at least in part by the elevated cyclin D1 and c-Jun expression caused by PAUF-induced β-catenin. Because the PAUF-β-catenin signaling axis may be a crucial therapeutic target in pancreatic tumors, we are currently screening small molecules that may block activation of PAUF-mediated β-catenin signaling and thereby retard or block pancreatic cancer growth.
Methods
Cell cultures and transfection
Panc-1 cells stably expressing vector (Panc-Vec) or PAUF (Panc-PAUF), BxPC3 cells stably expressing control shRNA (Bx-ConshRNA) or PAUF shRNA (Bx-PAUFshRNA), Wnt3a-secreting L cells, and HEK293 cells expressing Frizzled (HEK293-Fz) were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 1% penicillin and streptomycin, and G418 (500 µg/ml). HEK293-Fz cells were plated at 5 × 105 cells per 60-mm-diameter plate at 24 h before transfection and the cells were then transfected with 2 µg of DNA using lipofectamine 2000 (Invitrogen, Carlsbad, CA).
Reagents and antibodies
Wortmannin, LiCl, H-89, Bt2-cAMP, PD098059, SP600125 and SB202190 were purchased from Calbiochem (San Diego, CA). Hexachlorophene was purchased from Sigma (St. Louis, MO). For immunoblotting, anti-GSK-3β, anti-c-Jun, anti-Ras and β-tubulin antibodies were obtained from Santa Cruz Biotech (Santa Cruz, CA). Anti-Akt, anti-phospho-Akt (Ser-473), anti-phospho-GSK-3β (Ser-9), anti-ERK1/2, anti-phospho-ERK (Thr-202/Tyr-204), anti-JNK, anti-phospho-JNK(Thr-183/Tyr-185), anti-p38, anti-phospho-p38 (Thr-180/Tyr-182), anti-β-catenin and anti-phospho-β-catenin (Ser-33/37/Thr-41, Ser-45, or Ser-675) antibodies were acquired from Cell Signaling Biotech (Danvers, MA). Glutathione S-transferase (GST)-Raf-RBD fusion protein was obtained from Cytoskeleton (Denver, CO) to capture active Ras. For Wnt3a treatment, Wnt3a was used from culture supernatants from Wnt3a-secreting L cells.
Western blotting assay
Cells were harvested and lysed with lysis buffer (0.15 M NaCl, 1% Nonidet P-40, 50 mM Tris [pH7.5]) containing 0.1 mM Na2VO3, 1 mM NaF, and protease inhibitors (Sigma-Aldrich, St. Louis, MO). For immunoblotting, proteins from whole cell lysates were resolved by 10% or 12% SDS-PAGE and transferred to nitrocellulose membranes. Primary antibodies were used at 1:1,000 or 1:2,000 dilutions, and secondary HRP-conjugated antibodies (Santa Cruz) were used at 1:2,000 dilution in 5% nonfat dry milk. After final washing, nitrocellulose membranes were exposed using chemiluminescence assays (GE-Amersham, Piscataway, NJ).
Luciferase reporter assay
HEK 293 T cells were transfected with TOP Flash- or FOP Flash-vectors. To normalize transfection efficiency, a pGK-βgal vector that expresses β-galactosidase from a phosphoglucokinase promoter was included in the transfection mixture. At 48 h post-transfection, cells were washed with cold PBS and lysed in lysis solution (25 mM Tris [pH7.8], 2 mM EDTA, 2 mM DTT, 10% glycerol, and 1% Triton-X100). Luciferase activity was measured with a luminometer by using a luciferase kit (Promega, Madison, WI).
Reverse transcription-polymerase chain reaction (RT-PCR)
Total RNA was extracted from cells using RNeasy protect cell mini kit (Qiagen, Valencia, CA) in accordance with the manufacturer's instructions. Three micrograms of total RNA was converted to cDNA using Superscript II reverse transcriptase (Invitrogen), and PCR was performed using specific primers as described elsewhere (Jang et al., 2005; Choi et al., 2008). The cDNAs of each sample were diluted and PCR was run at the optimized cycle number. Beta-actin mRNA was measured as an internal standard. The relative ratio of β-catenin/β-actin level was expressed after measurement of the band intensity using Multi Gauge Ver. 2.1 (Fuji Photo Film, Japan).
Active Ras capture assay
To capture GTP-bound Ras from cell lysates, a GST-Raf-RBD fusion protein was used as described elsewhere (Ayroldi et al., 2002). The cells were lysed with lysis buffer and supernatants were incubated with glutathione-agarose on ice for 30 min to exclude nonspecific binding. After centrifugation, the supernatant was harvested, followed by adding GST-Raf-RBD (Cytoskeleton). The solution was incubated on ice for 1 h followed by adding glutathione-agarose to capture active Ras, which can bind to GST-Raf-RBD. To detect the captured Ras, immunoblotting was performed with anti-Ras antibody.
Statistical analysis
Data were presented as mean ± standard error. The Student t test was used for statistical analysis, with a P value < 0.05 defined as significance.
Abbreviations
- CK1:
-
casein kinase 1
- GSK-3β:
-
glycogen synthase kinase-3β
- PAUF:
-
pancreatic adenocarcinoma up-regulated factor
- PKA:
-
protein kinase A
- sh-RNA:
-
short hairpin-RNA
- TCF/LEF:
-
T cell factor/lymphocyte enhancer factor
References
Aberle H, Bauer A, Stappert J, Kispert A, Kemler R . Beta-catenin is a target for the ubiquitin-proteasome pathway . EMBO J 1997 ; 16 : 3797 - 3804
Ayroldi E, Zollo O, Macchiarulo A, Di Marco B, Marchetti C, Riccardi C . Glucocorticoid-induced leucine zipper inhibits the Raf-extracellular signal-regulated kinase pathway by binding to Raf-1 . Mol Cell Biol 2002 ; 22 : 7929 - 7941
Castellone MD, De Falco V, Rao DM, Bellelli R, Muthu M, Basolo F, Fusco A, Gutkind JS, Santoro M . The beta-catenin axis integrates multiple signals downstream from RET/papillary thyroid carcinoma leading to cell proliferation . Cancer Res 2009 ; 69 : 1867 - 1876
Choi SH, Jin SE, Lee MK, Lim SJ, Park JS, Kim BG, Ahn WS, Kim CK . Novel cationic solid lipid nanoparticles enhanced p53 gene transfer to lung cancer cells . Eur J Pharm Biopharm 2008 ; 68 : 545 - 554
Ge X, Jin Q, Zhang F, Yan T, Zhai Q . PCAF acetylates β-catenin and improves its stability . Mol Biol Cell 2009 ; 20 : 419 - 427
Ghaneh P, Costello E, Neoptolemos JP . Biology and management of pancreatic cancer . Gut 2007 ; 56 : 1134 - 1152
Giles RH, van Es JH, Clevers H . Caught up in a Wnt storm: Wnt signaling in cancer . Biochim Biophys Acta 2003 ; 1653 : 1 - 24
Hawes RH, Xiong Q, Waxman I, Chang KJ, Evans DB, Abbruzzese JL . A multispecialty approach to the diagnosis and management of pancreatic cancer . Am J Gastroenterol 2000 ; 95 : 17 - 31
He TC, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT, Morin PJ, Vogelstein B, Kinzler KW . Identification of c-MYC as a target of the APC pathway . Science 1998 ; 281 : 1509 - 1512
He TC, Chan TA, Vogelstein B, Kinzler KW . PPARdelta is an APC-regulated target of nonsteroidal anti-inflammatory drugs . Cell 1999 ; 99 : 335 - 345
Heiser PW, Lau J, Taketo MM, Herrera PL, Hebrok M . Stabilization of beta-catenin impacts pancreas growth . Development 2006 ; 133 : 2023 - 2032
Heiser PW, Cano DA, Landsman L, Kim GE, Kench JG, Klimstra DS, Taketo MM, Biankin AV, Hebrok M . Stabilization of beta-catenin induces pancreas tumor formation . Gastroenterology 2008 ; 135 : 1288 - 1300
Hino S, Tanji C, Nakayama KI, Kikuchi A . Phosphorylation of beta-catenin by cyclic AMP-dependent protein kinase stabilizes beta-catenin through inhibition of its ubiquitination . Mol Cell Biol 2005 ; 25 : 9063 - 9072
Iacobuzio-Donahue CA, Ashfaq R, Maitra A, Adsay NV, Shen-Ong GL, Berg K, Hollingsworth MA, Cameron JL, Yeo CJ, Kern SE, Goggins M, Hruban RH . Highly expressed genes in pancreatic ductal adenocarcinomas: a comprehensive characterization and comparison of the transcription profiles obtained from three major technologies . Cancer Res 2003a ; 63 : 8614 - 8622
Iacobuzio-Donahue CA, Maitra A, Olsen M, Lowe AW, van Heek NT, Rosty C, Walter K, Sato N, Parker A, Ashfaq R, Jaffee E, Ryu B, Jones J, Eshleman JR, Yeo CJ, Cameron JL, Kern SE, Hruban RH, Brown PO, Goggins M . Exploration of global gene expression patterns in pancreatic adenocarcinoma using cDNA microarrays . Am J Pathol 2003b ; 162 : 1151 - 1162
Jang KL, Shackelford J, Seo SY, Pagano JS . Up-regulation of beta-catenin by a viral oncogene correlates with inhibition of the seven in absentia homolog 1 in B lymphoma cells . Proc Natl Acad Sci USA 2005 ; 102 : 18431 - 18436
Kim SA, Lee Y, Jung DE, Park KH, Park JY, Gang J, Jeon SB, Park EC, Kim YG, Lee B, Liu Q, Zeng W, Yeramilli S, Lee S, Koh SS, Song SY . Pancreatic adenocarcinoma up-regulated factor (PAUF), a novel up-regulated secretory protein in pancreatic ductal adenocarcinoma . Cancer Sci 2009 ; 100 : 828 - 836
Kim SE, Choi KY . EGF receptor is involved in WNT3a-mediated proliferation and motility of NIH3T3 cells via ERK pathway activation . Cell Signal 2007 ; 19 : 1554 - 1564
Lee Y, Kim SJ, Park HD, Park EH, Huang SM, Jeon SB, Kim JM, Lim DS, Koh SS . PAUF functions in the metastasis of human pancreatic cancer cells and upregulates CXCR4 expression . Oncogene 2010 ; 29 : 56 - 67
Liu C, Li Y, Semenov M, Han C, Baeg GH, Tan Y, Zhang Z, Lin X, He X . Control of beta-catenin phosphorylation/degradation by a dual-kinase mechanism . Cell 2002 ; 108 : 837 - 847
Maitra A, Hruban RH . Pancreatic cancer . Annu Rev Pathol 2008 ; 3 : 157 - 188
Min HJ, Cho IR, Srisuttee R, Park EH, Cho DH, Ahn JH, Lee IS, Johnston RN, Oh S, Chung YH . Hexachlorophene suppresses beta-catenin expression by up-regulation of Siah-1 in EBV-infected B lymphoma cells . Cancer Lett 2009 ; 276 : 136 - 142
Nakagiri S, Murakami A, Takada S, Akiyama T, Yonehara S . Viral FLIP enhances Wnt signaling downstream of stabilized beta-catenin, leading to control of cell growth . Mol Cell Biol 2005 ; 25 : 9249 - 9258
Polakis P . Wnt signaling and cancer . Genes Dev 2000 ; 14 : 1837 - 1851
Rubinfeld B, Albert I, Porfiri E, Fiol C, Munemitsu S, Polakis P . Binding of GSK3beta to the APC-beta-catenin complex and regulation of complex assembly . Science 1996 ; 272 : 1023 - 1026
Saadeddin A, Babaei-Jadidi R, Spencer-Dene B, Nateri AS . The links between transcription, beta-catenin/JNK signaling, and carcinogenesis . Mol Cancer Res 2009 ; 7 : 1189 - 1196
Taurin S, Sandbo N, Qin Y, Browning D, Dulin NO . Phosphorylation of beta-catenin by cyclic AMP-dependent protein kinase . J Biol Chem 2006 ; 281 : 9971 - 9976
Tetsu O, McCormick F . Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells . Nature 1999 ; 398 : 422 - 426
van Noort M, Meeldijk J, van der Zee R, Destree O, Clevers H . Wnt signaling controls the phosphorylation status of beta-catenin . J Biol Chem 2002 ; 277 : 17901 - 17905
Wu H, Symes K, Seldin DC, Dominguez I . Threonine 393 of beta-catenin regulates interaction with Axin . J Cell Biochem 2009 ; 108 : 52 - 63
Acknowledgements
The study was supported for two years by Pusan National University Research Grant.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Cho, IR., Koh, S., Min, HJ. et al. Pancreatic adenocarcinoma up-regulated factor (PAUF) enhances the expression of β-catenin, leading to a rapid proliferation of pancreatic cells. Exp Mol Med 43, 82–90 (2011). https://doi.org/10.3858/emm.2011.43.2.010
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.3858/emm.2011.43.2.010
Keywords
This article is cited by
-
Pancreatic cancer induces muscle wasting by promoting the release of pancreatic adenocarcinoma upregulated factor
Experimental & Molecular Medicine (2021)
-
BCL9L expression in pancreatic neoplasia with a focus on SPN: a possible explanation for the enigma of the benign neoplasia
BMC Cancer (2016)
-
Silencing pancreatic adenocarcinoma upregulated factor (PAUF) increases the sensitivity of pancreatic cancer cells to gemcitabine
Tumor Biology (2016)
-
CXCR4 promotes GSK3β expression in pancreatic cancer cells via the Akt pathway
International Journal of Clinical Oncology (2015)
-
Effects of cyclin-dependent kinase 8 specific siRNA on the proliferation and apoptosis of colon cancer cells
Journal of Experimental & Clinical Cancer Research (2011)