Abstract
Cowden syndrome (CS) is an inherited autosomal dominant disorder associated with germline pathogenic variants of the PTEN tumor suppressor gene. Its phenotypical expression is highly variable and the existence of patients with a CS suggestive phenotype without pathogenic PTEN variant may be related to genetic heterogeneity. In order to explore this hypothesis through the detection of potentially deleterious variants enabling us to identify a new candidate gene, we performed whole-exome sequencing (WES) in a series of 22 CS patients without detectable PTEN pathogenic variant using conventional methods for mutation screening. We failed to identify a novel candidate gene, but interestingly in two patients WES revealed the presence of two distinct, previously undescribed Alu insertions with the same break points in exon 5. These insertions were not found in a series of 35 breast carcinomas that showed a loss of PTEN expression without a detectable alteration of this gene. This study reveals the presence of a PTEN Alu insertion hotspot involved in CS, and suggests that undetected PTEN pathogenic variants could contribute to CS.
Similar content being viewed by others
Introduction
Cowden syndrome (CS) (MIM 158350) is a rare inherited autosomal dominant disorder with high penetrance associated with germline pathogenic variants of the PTEN tumor suppressor gene.1 It is characterized by macrocephaly, hamartomas of multiple organs and an increased risk of neoplasia, with a particularly elevated risk for breast, thyroid and endometrial cancer.2, 3 Its clinical expression is highly variable and germline PTEN pathogenic variants have been described in various phenotypes including Bannayan–Riley–Ruvalcaba syndrome, Lhermitte–Duclos syndrome, juvenile polyposis and autism – macrocephaly syndrome. This variability has led to the use of an umbrella term, ‘PHTS’ for ‘PTEN hamartoma tumor syndrome’, that defines clinical presentations linked to germline PTEN pathogenic variants.4 PTEN encodes a phosphatase that negatively regulates the PIK3CA/AKT pathways. It acts as a tumor suppressor gene involved in a large proportion of malignant tumors including breast carcinoma. The broadening of the phenotypic spectrum in PHTS has been associated with a decrease in the PTEN pathogenic variant detection rate in a clinical diagnostic setting. It is currently accepted that ~20% of patients with a typical phenotype of CS remain unexplained following PTEN molecular analysis, suggesting a possible genetic heterogeneity for this disease.4 In order to explore this hypothesis, we performed whole-exome sequencing in a series of 22 PTEN mutation-negative CS patients.
Materials and methods
Patients and samples
The 22 index cases met the clinical diagnostic criteria for familial or sporadic CS according to the 2006 International Cowden Consortium with the exception of three patients with different PHTS. PTEN Cleveland Clinic5 and PTEN Predict6 diagnostic scores, which give the probability of detecting a PTEN pathogenic variant based on clinical features, were calculated for each patient and are provided in Supplementary Table 1. All participants had previously been tested negative for germline PTEN pathogenic variants using a combination of enhanced mismatch mutation analysis (EMMA, Fluigent, Paris, France),7 Sanger sequencing and quantitative multiplex PCR of short fragments (QMPSF).8 All patients agreed to the use of their samples for germline genetic analysis in compliance with the French law (law n°2004-800).
In addition, 35 breast carcinoma samples previously investigated for the involvement of PTEN using both immunohistochemical (IHC) and genetic analysis,9 and were selected based on their complete loss of PTEN IHC expression and absence of PTEN genetic alteration (point mutation or large genomic rearrangement). Their principal clinicopathological characteristics are provided in Supplementary Table 2.
Exome sequencing
Exome capture using SureSelect Human All Exon Kit (V5; Agilent, Santa Clara, CA, USA), and paired-end sequencing on an Illumina HiSeq 2000 system (Illumina, San Diego, CA, USA) were performed by the IntegraGen society (IntegraGen SA, Evry, France) using DNA extracted from leukocyte samples for all index cases. Bioinformatic analysis was based on the Illumina pipeline CASAVA1.8 using ELANDv2e alignment algorithms and the hg19 reference human genome. Annotation of genetic variants was performed using an IntegraGen custom pipeline. Overall sequencing coverage of the captured regions was 95% and 87% for a × 10 and × 25 depth of coverage, respectively, resulting in a mean sequencing depth of × 70 per base.
Sanger sequencing
DNA was amplified by PCR using QIAGEN Multiplex PCR Kit or AmpliTaq Gold Kit (Life Technologies, Paisley, UK). Break points-specific primers and conditions of PCR are provided in Supplementary Table 3. Sanger sequencing was performed using the ABI 3130xl DNA Analyzer (Life Technologies).
Alu insertion-dedicated PCR
In order to reveal the presence of the PTEN allele carrying the exon 5 Alu insertion by PCR, we designed an optimized dedicated PCR using the QIAGEN Multiplex PCR Kit. The specific primers and conditions are provided in Supplementary Table 3.
RT-PCR
Leukocytes RNA (125 ng) were reverse transcribed, then amplified using a specific PCR that amplified both wild-type and mutated PTEN transcripts. Sanger sequencing was performed using the ABI 3130xl DNA Analyzer (Life Technologies). Specific primers and PCR conditions are provided in Supplementary Table 4.
Results
Identification of PTEN Alu insertions in exome data
We were unable to identify a new candidate gene for Cowden disease in the 20 exomes analyzed by looking for loss-of-function mutations affecting the same gene in at least two families. Unexpectedly however, a PTEN exonic variant was reported in the variant calling format (.vcf) files (Supplementary Table 5) for two unrelated patients (patients 1998101 and 2001012) with the highest diagnostic scores (Supplementary Table 1). Their phenotypic characteristics are summarized in Table 1. The two variants consist of large insertions sharing the same break point at the end of exon 5 detected with low variant read percentages (4% for patient 1998101 and 8.5% for patient 2001012). Using the Alamut software to visualize aligned reads on the PTEN reference sequence, the identified break point was observed for both patients, as well as many unpaired reads whose partner read did not align to PTEN (Supplementary Figure 1). Investigation of these partner reads using the IGV software (Broad Institute, Cambridge, MA, USA) for both patients allowed us to complete the inserted sequences given in the.vcf files. For patient 1998101, these sequences aligned to an intergenic region in 4q22.3 between the genes PDHA2 and STPG2 (Supplementary Table 5). For patient 2001012, they aligned to an intronic region of the MB21D1 gene (Supplementary Table 5). These regions were identified as Alu elements by the RepeatMasker Element (University of California, Santa Cruz Genome Browser) and alignment of the two inserted sequences in Repbase Giri,10 showed that one was an AluYa5 element (patient 1998101), and the other a 5′ truncated AluYb8 element (patient 2001012). Both Alu elements were inserted in an antisense orientation with respect to the PTEN reading frame.
Analysis of insertion sites
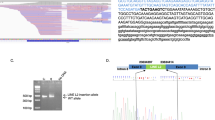
Alu insertion-specific primers, in combination with PTEN exon 5-specific primers (Supplementary Table 3), were used to specifically amplify and sequence the mutant alleles. The first break point, located in a highly conserved TTTT / AA motif, known as the LINE1 endonuclease consensus cleavage site11 corresponds to the boundary of the 3′ portion (poly T tail) of the Alu element (Figures 1a and b). The second break point corresponds to the junction between the 5′ portion of the Alu sequence and an extremity of the PTEN sequence involved in a short duplication of 17 base pairs flanking the inserted Alu and known as ‘targeted site duplication’ (TSD). Both the break points and the TSD are identical for the two patients (Figures 1a and b; Supplementary Figure 1). The nomenclature for these PTEN variants according to the Human Genome Variation Society using the PTEN cDNA sequence for which nucleotide 1 is the A of the ATG translation initiation codon and exons are numbered according to NG_007466.2 is as follows:
Alu insertions in PTEN exon 5 in two Cowden disease patients. Schematic representation of the Alu elements inserted into exon 5 of PTEN and Sanger sequencing of break points from mutant alleles for patient 1 (a) and patient 2 (b). The insertion was in an antisense orientation compared to the reference sequence. (c) Gel electrophoresis of PTEN exon 5 PCR products from two control individuals (C1 and C2), and patients 1998101 (P1) and 2001012 (P2) harboring a full-length Alu element (P1) and a truncated Alu element (P2). The additional bands are probably derived from heteroduplexes between the wild-type and mutant alleles, neither of which are present in the controls. TSD, target site duplication; W, water control.
-NM_000314.4:c.[437_438insNC_000004.11:g.97897210_97897530;ins421_437], which can be simplified as c.437_438insAluYa5 for patient 1998101.
-NM_000314.4:c.[437_438insNC_000006.11:g.74156323_74156488;ins421_437], which can be simplified as c.437-438insAluYb8 for patient 2001012.
These two PTEN mutations have been submitted to the LOVD database (www.lovd.nl/PTEN) under the accession numbers: ID 00081841 and ID00081842, respectively, for patient 1998101 and 2001012.
Transcriptional consequences of the PTEN Alu insertions
The genetic variant observed in patient 1998101 consists of a frameshift insertion of 338 nucleotides in exon 5 of PTEN leading to a premature stop codon following 87 modified amino acids [p.(Leu146Phefs*88)]. For patient 2001012, the variant consists of an in frame insertion of 183 nucleotides leading to a stop codon within the inserted sequence following 50 modified amino acids [p.(Leu146_Val403delins50*)]. Lymphocytic RNA was available for patient 2001012, thus we were able to characterize the mutated allele at the RNA level using RT-PCR covering a region between exons 2 and 7 of PTEN. Visualization of the RT-PCR product by gel electrophoresis did not reveal a product larger than the wild type, but revealed the presence of a shorter product, absent in controls, which corresponded to a transcript with full skipping of exon 5 (Supplementary Figure 2). Because PTEN exon 5 is not in frame, this mutant allele also leads to a frameshift and a premature stop codon [p.(Val85Glyfs*15)]. It was not possible to evaluate the allelic ratio between the wild-type and mutated alleles at the RNA level, as no single-nucleotide polymorphism allowing the distinction of the two transcribed alleles was present in any of the PTEN exons (including 5′ and 3′ untranslated regions) for this patient.
Cosegregation of the mutated allele and the disease in family 2001012
Family 2001012 harbored three affected members, the propositus and, her son and her daughter which both present macrocephaly, mucocutaneous features of the disease and thyroid lesions (Supplementary Figure 3A). Dedicated PCR amplifying both wild-type allele and the exon 5 Alu insertion (Figure 1c) revealed that all three affected members of this family carried the mutated allele (Supplementary Figure 3B). The identical electrophoresis profiles for the three patients confirm that it is an inherited mutation in the three carriers in this family and not a mosaic mutation, even for the propositus.
Cosegregation analysis was not feasible because there was only one known affected individual in family 1998101 for which only limited information was available.
Potential involvement of PTEN exon 5 Alu insertion in breast cancer
PTEN is a tumor suppressor gene involved in many non-hereditary malignant tumors including breast cancer. A loss of expression of this gene, revealed by immunohistochemistry is observed in around 20% of breast carcinoma. However, genetic alterations such as point mutations or intragenic rearrangements are only observed for a minority of cases (5% of breast carcinoma).9 As an insertion of a LINE1 HS element has recently been described within exon 6 of PTEN in an endometrial carcinoma,12 we postulated that the exon 5 Alu insertion could explain some breast cancer cases with a loss of PTEN expression without an associated detectable mutation. In order to explore this hypothesis, we screened 35 previously selected breast cancers showing such characteristics9 using the PTEN exon 5 Alu insertion-dedicated PCR. No Alu insertion was detected at the insertion hotspot in any of these tumors (data not shown), indicating that this mutation is unlikely to explain the low PTEN expression that has previously been reported in these tumors.
Discussion
We report two CS patients carrying distinct Alu insertions with the same break points in the PTEN gene that were previously undetected using conventional methods. To our knowledge, such alterations have not been reported in CS to date. This is probably because PTEN analysis has, until recently, mainly been performed using PCR-based methods for which standard conditions lead to only wild-type and not Alu insertion allele amplification due to its increased size and to the poly (T) tail of the Alu element. Interestingly, for both patients the initial EMMA screening showed slightly abnormal exon 5 profiles with a decreased intensity but QMPSF and Sanger sequencing of exon 5 failed to detect any alteration. Next-generation sequencing (NGS) is able to detect this type of variant if the bioinformatic pipeline retains unpaired reads and does not eliminate reads with segmental alignment.13 In the two cases reported here the number of reads linked to the mutated allele was very low (4 and 8.5%). This is unexpectedly low for a heterozygote inherited mutation and is probably due to the lack of capture of the inserted sequence during library preparation and the elimination of most of the chimaeric reads during the alignment process. We recommend that low-frequency variants observed in NGS results should be taken into consideration in order to detect large insertions and structural variants.
The exact consequences of these Alu insertions within PTEN exon 5 are not clearly defined. It is most likely that transcripts retaining the inserted sequences are eliminated by nonsense-mediated decay (NMD) because of the creation of a premature stop codon regardless of the inserted sequence reading frame. However, as shown for patient 2001012 (Supplementary Figure 2), these mutations can also lead to abnormal splicing, either complete or incomplete. In the case documented here, skipping of exon 5 also leads to a premature stop codon that probably activates NMD as suggested by the low intensity of the skipped exon 5 allele in RT-PCR gel electrophoresis (Supplementary Figure 2A). This loss of a PTEN allele is clearly involved in the typical Cowden phenotype shown by these patients.
The fact that two distinct Alu elements were inserted with identical break points indicates the presence of a retrotransposition hotspot at this locus. It is therefore expected that other CS patients may carry the same type of pathogenic variant as is seen in other human diseases, either resulting from a founder mutation as for the BRCA2 c.156-157insAlu mutation14 or due to an insertional hotspot as for the NF1 gene.15 We therefore recommend that molecular analysis of the PTEN gene for PHTS includes systematic screening for Alu insertions within exon 5, using either a specific PCR or an adapted pipeline for NGS data analysis.
Both of the patients described showed high PTEN-Predict and PTEN Cleveland Clinic Scores (Supplementary Table 1), and have a very characteristic Cowden disease. However, it is interesting to note that, at 43 and 62 years old, respectively, they have not presented any malignant tumor, which is rarely observed in CS (cumulative risk at 60 years: 75%).2 This observation in association with the non-involvement of the exon 5 Alu insertion in breast carcinomas with PTEN loss of expression raises the question of an absence of tumorigenicity of these specific mutations as it has been reported for other PTEN mutations in mouse models.16
The large phenotypic variation in PHTS has led to suspect the involvement of PTEN in a large variety of clinical conditions, thus increasing the number of suspected PHTS conditions without a molecular genetic cause. The involvement of genes other than PTEN has been described in such conditions, leading to the implication of the SDH genes,17 PIK3CA, AKT118 or more recently SEC23B19 in CS. This genetic heterogeneity has not been confirmed however and has even been contested.20 The detection of previously unidentified PTEN deleterious variants, specifically in patients with a highly suggestive phenotype of CS justifies the reevaluation of the contribution of allelic heterogeneity rather than genetic heterogeneity to explain such phenotypes. Deep intronic mutations or PTEN regulatory element alterations may also represent currently undetected deleterious variants in CS.
References
Liaw D, Marsh D, Li J : Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nat Genet 1997; 16: 64–67.
Bubien V, Bonnet F, Brouste V et al: High cumulative risks of cancer in patients with PTEN hamartoma tumour syndrome. J Med Genet 2013; 50: 255–263.
Mester J, Eng C : Cowden syndrome: recognizing and managing a not-so-rare hereditary cancer syndrome: Cowden syndrome. J Surg Oncol 2015; 111: 125–130.
Pilarski R, Eng C : Will the real Cowden syndrome please stand up (again)? Expanding mutational and clinical spectra of the PTEN hamartoma tumour syndrome. J Med Genet 2004; 41: 323–326.
Tan M-H, Mester J, Peterson C et al: A clinical scoring system for selection of patients for PTEN mutation testing is proposed on the basis of a prospective study of 3042 probands. Am J Hum Genet 2011; 88: 42–56.
Pilarski R, Stephens JA, Noss R, Fisher JL, Prior TW : Predicting PTEN mutations: an evaluation of Cowden syndrome and Bannayan-Riley-Ruvalcaba syndrome clinical features. J Med Genet 2011; 48: 505–512.
Houdayer C, Moncoutier V : Enhanced mismatch mutation analysis: simultaneous detection of point mutations and large scale rearrangements by capillary electrophoresis, application to BRCA1 and BRCA2. Methods Mol Biol 2010; 653: 147–180.
Casilli F, Di Rocco ZC, Gad S et al: Rapid detection of novel BRCA1 rearrangements in high-risk breast-ovarian cancer families using multiplex PCR of short fluorescent fragments. Hum Mutat 2002; 20: 218–226.
Jones N, Bonnet F, Sfar S et al: Comprehensive analysis of PTEN status in breast carcinomas: PTEN status in breast cancer. Int J Cancer 2013; 133: 323–334.
Jurka J : Repbase update: a database and an electronic journal of repetitive elements. Trends Genet 2000; 16: 418–420.
Chen J-M, Férec C, Cooper DN : Mechanism of Alu integration into the human genome. Genomic Med 2007; 1: 9–17.
Helman E, Lawrence MS, Stewart C, Sougnez C, Getz G, Meyerson M : Somatic retrotransposition in human cancer revealed by whole-genome and exome sequencing. Genome Res 2014; 24: 1053–1063.
Samuels DC, Han L, Li J et al: Finding the lost treasures in exome sequencing data. Trends Genet 2013; 29: 593–599.
Peixoto A, Santos C, Pinheiro M et al: International distribution and age estimation of the Portuguese BRCA2 c.156_157insAlu founder mutation. Breast Cancer Res Treat 2010; 127: 671–679.
Wimmer K, Callens T, Wernstedt A, Messiaen L : The NF1 gene contains hotspots for L1 endonuclease-dependent de novo insertion. PLoS Genet 2011; 7: e1002371.
Wang H, Karikomi M, Naidu S et al: Allele-specific tumor spectrum in Pten knockin mice. Proc Natl Acad Sci USA 2010; 107: 5142–5147.
Ni Y, Zbuk KM, Sadler T et al: Germline mutations and variants in the succinate dehydrogenase genes in Cowden and Cowden-like syndromes. Am J Hum Genet 2016; 83: 261–268.
Orloff MS, He X, Peterson C et al: Germline PIK3CA and AKT1 mutations in Cowden and Cowden-like syndromes. Am J Hum Genet 2016; 92: 76–80.
Yehia L, Niazi F, Ni Y et al: Germline heterozygous variants in SEC23B are associated with Cowden syndrome and enriched in apparently sporadic thyroid cancer. Am J Hum Genet 2016; 97: 661–676.
Bayley J-P : Succinate dehydrogenase gene variants and their role in Cowden syndrome. Am J Hum Genet 2016; 88: 674–675.
Acknowledgements
We are grateful to the patients and their families. This work has been supported by the ‘Fondation ARC pour la recherche sur le cancer’ (LC) and ‘le Lions club de Bergerac’. We would like to thank Dr Ravi Nookala of Institut Bergonié for the medical writer service and Bernadette Gastaldello for technical assistance.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Crivelli, L., Bubien, V., Jones, N. et al. Insertion of Alu elements at a PTEN hotspot in Cowden syndrome. Eur J Hum Genet 25, 1087–1091 (2017). https://doi.org/10.1038/ejhg.2017.81
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2017.81
This article is cited by
-
Considerations on diagnosis and surveillance measures of PTEN hamartoma tumor syndrome: clinical and genetic study in a series of Spanish patients
Orphanet Journal of Rare Diseases (2022)
-
A Variety of Alu-Mediated Copy Number Variations Can Underlie IL-12Rβ1 Deficiency
Journal of Clinical Immunology (2018)