Abstract
Spinocerebellar ataxias (SCA) comprise a heterogeneous group of inherited neurological disorders characterized by a range of symptoms from both cerebellar and extra cerebellar structures. We investigated the cause of autosomal recessive, congenital SCA in six affected family members from a large consanguineous family. Using whole-exome sequencing, we identified a homozygous ITPR1 missense variant [c.5360T>C; p.(L1787P)] segregating in all affected individuals. Heterozygous carriers were asymptomatic despite cerebellar hypoplasia. Variants in the ITPTR1 gene have previously been associated exclusively with autosomal dominant SCA15 and SCA29 with slow or no progression. The L1787 residue is highly conserved and the leucine to proline substitution has a predicted destabilizing effect on the protein structure. Additionally, the L1787P variant is located in a domain separated from previously described and dominant-acting missense variants consistent with a distinct effect on IP3R1 tetramer structure and function. Taken together, we show for the first time that a biallelic ITPR1 missense variant may cause an autosomal recessive and infantile onset SCA29, albeit with subclinical cerebellar hypoplasia in carriers. Our findings add to the genetic complexity of SCA29 and broaden the correlations between ITPR1 variants and their clinical expression.
Similar content being viewed by others
Introduction
Spinocerebellar ataxias (SCA) are a heterogeneous group of neurodegenerative disorders characterized by loss of motor control.1, 2 Typical clinical features are ataxia with loss of gait and limb coordination, dysarthria, sometimes accompanied with altered ocular movements. To date more than 40 different SCA types are described with an extensive genetic heterogeneity and with all modes of transmissions represented.3
The ITPR1 gene, encoding IP3R1, has to date been associated with a broad spectrum of phenotypes derived mainly from the cerebellum. In the central nervous system, IP3R1 is abundantly expressed and forms homo- or heterotetramers together with the homologous IP3R2 and IP3R3 that act as Ca2+ release channels in the endoplasmic reticulum.4, 5 Neurological characteristics associated with ITPR1 mutations were first modeled in mice and prior to the discovery in humans. Already 20 years ago the targeted ablation of murine Itpr1 was shown to cause early onset ataxia, dystonia, multiple abnormal movements and early death.6 Heterozygous deletions involving ITPR1 were later identified in autosomal dominant SCA15.7, 8, 9, 10, 11 Individuals with SCA15 have an adult onset, sometimes in late adulthood, usually with slow progression of neurological symptoms. Missense variants in ITPR1 are instead associated with autosomal dominant SCA29 characterized by onset of ataxia in infancy or childhood. Furthermore, SCA29 is often accompanied by delayed development and cognitive impairment, and some cases fulfill diagnostic criteria for ataxic cerebral palsy.12, 13, 14 Magnetic resonance imaging (MRI) of both clinical forms demonstrates cerebellar atrophy, often prominent and mainly affecting the vermis.9, 10, 13, 15 More recently, patients with cerebellar syndrome, intellectual disability and aniridia (Gillespie syndrome) were added to the ITPR1-associated phenotypes.14, 16 Taken together, the spectrum of ITPR1-associated phenotypes is intriguing. The heterozygous ITPR1 deletions in late onset SCA15 suggest haploinsufficiency as a disease mechanism. In contrast, cases with a congenital or infantile onset of SCA29 appear to be caused exclusively by monoallelic ITPR1 gene variants that alter the structure of IP3R1, presumably through a dominant negative effect.12, 14, 15, 16, 17, 18 To date, no autosomal recessive form of SCA29 has been attributed to ITPR1 variants.
We present herein a large consanguineous family segregating infantile onset autosomal recessive cerebellar syndrome and mild intellectual disability in six individuals. Symptoms were slowly progressive to stationary with close similarities to that of SCA29. A unique homozygous missense variant in ITPR1 was identified in the six affected members. Furthermore, we show that heterozygosity for the missense variant is associated with asymptomatic cerebellar hypoplasia. A video presentation of the study is available as Supplementary Information on European Journal of Human Genetics website.
Materials and methods
Patients
A five-generation consanguineous Pakistani family from the province of Punjab was identified segregating autosomal recessive congenital ataxia in two generations (Figure 1a). The family comprised two consanguineous loops with altogether six affected individuals. Fifteen family members were sampled and interviewed about their medical history. Eleven out of the 15 family members were examined by a neurologist. Two affected family members as well as two asymptomatic and obligate carriers were examined by MRI. Prior to start, ethical approval was obtained from the ethical committee of the National Institute of Biotechnology and Genetic Engineering (NIBGE), Faisalabad, Pakistan and according to the Declaration of Helsinki. Interviews, clinical examination and blood sampling of study participants were performed after informed written consent obtained from all participants or their legal guardians.
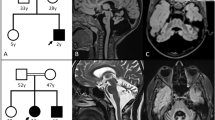
Genetic investigations of the family segregating SCA29. (a) Pedigree and genotypes of the consanguineous family segregating ITPR1 mutation c.5360T>C. The family comprises two consanguinity loops and six affected individuals. (b) Sequence chromatogram of genomic DNA showing part of exon 41 in the ITPR1 gene obtained from the healthy control V:4 (top), a heterozygous parent IV:2 (middle) and a homozygous affected individual V:1 (bottom). Arrows indicate the position of the c.5360T>C transition. (c) Relative position of the p.(Leu1787Pro) substitution in the modulatory domain of the IP3R protein and degree of conservation of the Leu1787 residue (shaded, bottom) across different species. For comparision, the region that spans known missense variants associated with autosomal dominant SCA is indicated and with a location toward the N-terminus, near the ligand transferase and ligand binding domains. Similarly, the region spanning missense variants associated with Gillespie syndrome is shown toward the C-terminus, near the intracellular transmembrane domain and the calcium ion transport channel. COOH, C-terminus; NH2, N-terminus.
Whole-exome sequencing and data analysis
Genomic DNA was extracted from peripheral blood leukocytes. The mode of inheritance and the genetic heterogeneity in SCA prompted us to perform whole-exome sequencing using 50 ng of DNA from two affected individuals (ind. V:1 and V:2). The DNA was sheared using a Covaris instrument (Covaris Inc., Woburn, MA, USA). Fragment libraries were created from the sheared samples using AB Library Builder System (Life Technologies, Carlsbad, CA, USA) and size selected on the BluePippin instrument (Sage Science, Beverly, MA, USA). Target enrichment was performed using the Ion AmpliSeq (Life Technologies), according to the manufacturer’s protocols. Briefly exome capture was conducted by hybridizing the DNA libraries with biotinylated RNA baits for 24 h followed by extraction using streptavidin-coated magnetic beads. Captured DNA was amplified by emulsion PCR using the Ion OneTouch 2 system and the Ion PI Template OT2 200 Kit chemistry (Life Technologies), followed by enrichment using Ion OneTouch ES. Samples were loaded on an Ion PI chip and sequenced on the Ion Proton System using Ion PI Sequencing 200 Kit (200 bp read length, Life Technologies).
Alignment of reads to the human reference sequence (hg19 assembly) and variant detection was performed using v2.1 of the LifeScope Software (Life Technologies). SNPs and indel data were stored in an in-house exome database together with variant annotation information obtained from ANNOVAR and dbSNP135.19 Custom R scripts were used to identify potentially damaging variants that were shared between the patients while not present in any of the other ~1000 exomes in the in-house database.
Segregation analysis was performed by bidirectional Sanger sequencing (Applied Biosystems Big Dye Terminator v3.1 Cycle Sequencing Kit, Applied Biosystems, Carlsbad, CA, USA; Life Technologies) on a 3730xl DNA Analyzer (Applied Biosystems, Life Technologies). Sequencher software (Gene Codes Corporation, Ann Arbor, MI, USA) was used for analysis of Sanger sequencing results.
In silico predictions of the effect of gene variants were performed using PolyPhen-2, MutationTaster and PROVEAN.20, 21, 22 The possible effect of the variant on splicing was investigated using BDGP: Splice Site Prediction.23 Two-point LOD score calculations were made with the MLINK program of the LINKAGE package version 5.1.24
3D modeling and predictions of protein stability
The 3D structure of the Ip3r1 tetramer from rat was used as model. The residue L1827 in the rat Ip3r1 corresponds to the human ITPR1 residue L1787 (3JAV L1827; human NP_001093422.2 L1787) and was illustrated using Yasara (www.YASARA.org). The impact of the variant on protein stability was predicted using mCSM and the available published crystal structure of the Ip3r1 tetramer from rat (PDB ID: 3JAV).25, 26
Results
Clinical characterization
Eleven family members underwent neurological investigation including six affected individuals and five asymptomatic individuals. The clinical features are summarized in Table 1. All affected individuals had an onset with generalized tremor of head, arms and trunk diagnosed at a few months of age. Ataxia became evident as soon as the infants were able to sit. None of the affected individuals did ever walk and their lack of independent ambulation made them develop a quadrupedal gait. All six patients had a delayed psychomotor development from childhood consistent with mild intellectual disability. Dysarthria became evident in the first decade. Truncal ataxia, tremor and mild intellectual disability remained stationary without deterioration in adulthood. None of the affected individuals had any visual problems and eye investigation revealed normal iris without hypoplasia or other morphological abnormalities. Gaze-evoked nystagmus was present in three affected individuals with an onset in their second or third decade. Dysmetria was present in 4/6 cases. Hyperreflexia and postural action tremor was observed in 5/6 cases. Five clinically unaffected family members (ind. III:1, III:2, IV:1, IV:2 and V:4) were examined at ages 78, 80, 45, 48 and 18 years, respectively. All five individuals considered themselves as healthy without gait disturbances and this was confirmed by a neurologist.
Magnetic resonance imaging
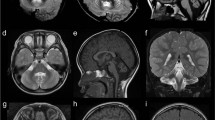
Two affected brothers (V:1 and V:2) and two asymptomatic individuals (IV:2 and V:4) were available for brain MRI. The MRI in the two clinically affected brothers revealed characteristic finding with cerebellar atrophy, most pronounced in the vermis (Figure 2). The midsagittal vermis areas of the affected brothers and asymptomatic carriers were measured twice and independently by two neuroradiologists. The mean vermis areas from the two measurements were 3.85 cm2 for ind. V:1 (4.1 and 3.6 cm2) and 5 cm2 for ind. V:2 (5.4 and 4.6 cm2), respectively. These values were compared to control values for the midsagittal vermis area calculated from MRI in 18 healthy individuals, 11 males and 7 women at age 19–49 years, showing a mean area of 15.1 cm2 (95% CI 13.9–16.3 cm2, range 11.2–18.1 cm2). No pontine abnormalities were detected. Furthermore, the mean vermis areas from two measurements in the two asymptomatic and heterozygous carriers were 10.25 cm2 for ind V:4 (11.1 and 9.4 cm2) and 10.65 cm2 for ind. IV:2 (11.9 and 9.4 cm2), respectively, and thus below both the 95% CI and range of vermis area for controls. Taken together, the MRI data indicated a marked atrophy in affected individuals as well as a distinguishable cerebellar hypoplasia in the two asymptomatic family members.
Brain imaging showing cerebellar abnormalities associated with homozygosity and heterozygosity for ITPR1 c.5360T>C, respectively. (a) MRI of individual V:2 homozygous for the c.5360T>C variant demonstrates marked atrophy of the cerebellar vermis (left panel, midsagittal area 5.4 cm2, black line) with sparing of the corpus callosum, brainstem and adjacent cerebral structures. Coronal section (right panel) illustrates volume reduction also of the cerebellar hemispheres, with widened cerebellar sulci. Sagittal and coronal T2-weighted spin echo (T2wSE) images. (b) MRI of the unaffected individual V:4 heterozygous for the c.5360T>C variant illustrating hypoplasia of the cerebellar vermis (left panel, midsagittal area 10.25 cm2, black line). Coronal section (right panel) illustrates slightly widened cerebellar sulci. Sagittal and coronal T2wSE images. Mean vermis midsagittal area from 18 healthy adults individuals was 15.1 cm2 (95% CI 13.9–16.3 cm2, range 11.2–18.1 cm2).
Whole-exome sequencing and genetic analyses
We filtered the WES data for shared homozygous or compound heterozygous variants in the two sequenced individuals. This revealed 14 homozygous and 7 compound heterozygous non-synonymous single-nucleotide variants, including a homozygous variant in the ITPR1 (c.5360T>C; transcript variant 1; NM_001099952.2) gene (Supplementary Table 1). We further filtered WES data for heterozygous variants to investigate the possible involvement of a dominant gene for ataxia. We performed in silico analysis of the 100 heterozygous variants shared by the two exomes from individuals V:1 and V:2. None of the genes containing shared heterozygous variants were previously associated with dominant ataxia when using the ataxia gene panels from the clinical diagnostic labs at the UK Genetic Testing Network (https://ukgtn.nhs.uk), Blueprint genetics (blueprintgenetics.com) and the ataxia exome panel from University of Chicago (dnatesting.uchicago.edu), comprising together in total 503 ataxia-related genes. Among the 21 recessive variants, 20 were excluded either by being present in homozygous form in the ExAc database without associated neurological features or by segregation analysis in our family (http://exac.broadinstitute.org).27 On the basis of the previous association of ITPR1 variants with SCA and the absence of other candidate variants we considered c.5360T>C as a plausible genetic cause for the disease. Sanger sequencing of the ITPR1 variant revealed that all six affected individuals are homozygous, while five asymptomatic family members are heterozygous, including individuals IV:2 and V:4 with cerebellar hypoplasia (Figures 1a and b), A LOD score of 3.94 (at θ=0.00) was obtained for the missense variant that is located in exon 41 in ITPR1 transcript 1 and alters an evolutionary conserved residue in a region encoding the modulatory domain of IP3R1 (Figure 1c). The resulting amino-acid substitution replaces a leucine for proline p.(L1787P) predicted to be damaging by MutationTaster (disease causing), PolyPhen-2 (probably damaging with a score of 1.0) and PROVEAN (deleterious with a score of 0.011). No effect of the variant on splicing was predicted using BDGP: Splice Site. The residue corresponding to human L1787 of IP3R1 is conserved in all 98 vertebrate species with sequence over this base according to the UCSC browser (Figure 1c; https://genome.ucsc.edu/; PhyloP score 7.58). Furthermore, the variant was excluded in 200 ethnically Pakistani control chromosomes and it is not present in the EVS data release (ESP6500) on the Exome Variant Server, NHLBI GO Exome Sequencing Project (ESP), Seattle, WA (http://evs.gs.washington.edu/EVS/) or the Exome Aggregation Consortium (ExAC) database, suggesting that the missense mutation is very rare. The c.5360T>C variant in ITPR1 has been reported to the ClinVar database with the accession number SCV000297811 (https://www.ncbi.nlm.nih.gov/clinvar/).
3D modeling
The 3D structure of the Ip3r1 tetramer in rat showed that residue L1827, corresponding to human residue L1787, is positioned in a peripheral domain, separated from known functional domains and the previously identified missense variants associated with either SCA or Gillespie syndrome (Figure 3). The L1787P substitution predicts a stability change of the protein (ΔΔG=−1.105 kcal/mol) that indicates a destabilizing effect similar to that for previously reported missense variants in IP3R1.16
Structure of the Ip3r1 tetramer in rat. (a) View from the top, showing the pore, and (b) from the side, showing the binding domains and the transmembrane domain (structure derived from PDB: 3JAV). The residue corresponding to amino acid L1787 in human IP3R1 is indicated in red. Position of missense variants associated with autosomal dominant Gillespie syndrome is mainly situated in the transmembrane domain and missense variants associated with SCA29 are situated toward the ligand binding domains of the tetramer. Dashed lines illustrate the plasma membrane of the endoplasmic reticulum (ER).
Discussion
We investigated a five-generation consanguineous family segregating autosomal recessive SCA in two sibships with altogether six affected individuals. Analysis of whole-exome sequencing data revealed a variant c.5360T>C in a conserved codon of ITPR1 as the unique remaining candidate after considering both dominant and recessive variants. All six affected individuals were homozygous for the variant and segregation analysis in 15 family members, including four obligate carriers, was consistent with autosomal recessive inheritance. Investigation by a neurologist confirmed a severe form of SCA in affected individuals with truncal ataxia, broad-based quadrupedal gait, dysarthria and intellectual disability. Family history revealed an onset in infancy with truncal ataxia and developmental delay but without evident progression of symptoms from the second decade. Cerebellar atrophy is usually pronounced in early onset SCA29 and this was indeed confirmed in two affected individuals showing marked reduction in cerebellar size, mainly of the vermis. In combination, the clinical and radiological findings of affected individuals were consistent with SCA29 and ataxic cerebral palsy.
The apparent autosomal recessive inheritance, supported by the segregation of the ITPR1 variant and clinical expression confined to homozygous family members, made us further investigate the heterozygous individuals. In total, five family members were confirmed as heterozygous carriers and they all denied any neurological or cognitive problems upon interviews. Investigation of two of carriers at 78 and 80 years of age, respectively, suggested that heterozygosity for the missense variant did not express clinically with age. The lack of symptoms in carriers was indeed supported by clinical examination performed by a neurologist on five heterozygous individuals that excluded mild cerebellar signs and symptoms (Table 1). To further exploit the possibility of subclinical signs of the disease, we performed MRI on two asymptomatic carriers for the missense variant. Surprisingly, both individuals showed cerebellar hypoplasia with vermis areas below the range of areas in normal, aged-matched individuals. Interestingly, in two previously reported families segregating late onset SCA15 and heterozygous ITPR1 deletions, mild gait disturbances and cerebellar atrophy were identified but unnoticed by the patients themselves.10 Together, this highlights the broad clinical spectrum associated with ITPR1 mutations, including subclinical features or subtle phenotypes.
Previous genotype–phenotype correlations have suggested that the pathogenic mechanism in SCA15 is mediated by haploinsufficiency, whereas the congenital or infantile onset SCA29 appears to be caused by monoallelic ITPR1 missense variants that alter the structure of IP3R1, presumably through a dominant negative effect.12, 18 Additionally, variants in the ITPR1 gene associated with the recessive form of Gillespie syndrome and with clinical similarities to SCA29, is mainly caused by LOF mutations. However, these mechanistic explanations are inconsistent with the observations in our family. Prediction of the structural destabilization of the missense variant on Ip3r1 in rat showed a structural destabilization of the protein when replacing leucine for proline at amino acid position 1827 that corresponds to residue 1787 in human IP3R1. Furthermore, in the 3D structure of the Ip3r1 tetramer, heterozygous missense variants associated with Gillespie syndrome are mainly located near the center of the channel, whereas the heterozygous missense variants associated with congenital forms of SCA are positioned within the cytoplasmic domains and closer to the InsP3-binding site.16 The p.(L1827) residue is positioned in a distinct cytoplasmic domain and further away from the ligand binding sites. This is also illustrated in the 2D model of the protein in which missense variants associated with SCA clustered toward the N-terminus, near the ligand transferase and ligand binding domains, whereas variants associated with autosomal dominant Gillespie syndrome cluster toward the C-terminus, near the intracellular transmembrane domain and the calcium ion transport channel.16 Thus, the position of the missense variant L1787P in a peripheral domain, separated from known functional domains and dominant acting variants, is consistent with a distinct effect on IP3R1 tetramer structure and function. Moreover, the fact that ITPR1 is among the most intolerant genes for missense variants with a z-score of 5.75 (ExAc database) provides indirect support for an effect of c.5360T>C.28 A possible mechanistic explanation is that the recessive inheritance in our family is caused by a partial loss of IP3R1 function in individuals who are heterozygous for c.5360T>C. This results in the relative loss of neuronal tissue in cerebellum and brainstem, however functionally compensated by remaining neurons and neural circuits. Individuals who are homozygous for the variant have an almost complete loss of IP3R1 tetramer function consistent with an early onset and a severe clinical expression.
The genotype–phenotype correlations in humans are to some extent consistent with that in mice: The Itpr1−/− model shows early onset ataxia, multiple abnormal movements and early death, whereas the heterozygous mouse (Itpr1wt/−) develop milder motor coordination at a later stage.29 Furthermore, a conditional deletion of the Itpr1 gene in Purkinje cells causes ataxia in mice and confirms the presence of IP3R1-dependent neural circuits in the cerebellum and the brainstem.30, 31
The ITPR1 gene product IP3R1 forms tetrameric intracellular Ca2+ release channel in the endoplasmic reticulum. In mammals, there are three highly homologous IP3Rs (IP3R1-3) and the three forms may assemble both as homo- and heterotetramers.5 In the central nervous system, IP3R1 is the major IP3R isoform and it is abundantly expressed in different brain regions including cerebellum, cerebral cortex and striatum.32 Previous studies have revealed an intricate balance between the three functionally redundant IP3R proteins that varies between tissues depending on their expression profiles.5 Consequently, perturbed function of one of the IP3Rs may result in a spectrum of consequences from no effect at all to an overt and tissue-specific disorder as shown for ITPR2.33 Thus, similar mechanisms may presumably lead to the phenotypes associated with ITPR1 mutations because of variable reductions in levels of functional homo- and heterotetramers in specific brain structures. Such a model is consistent with the relatively high expression levels of the IP3R1 isoform in cerebellum and cortex together with ataxia and cognitive impairment associated with ITPR1 variants. However, the precise mechanisms caused by the ITPR1 variant reported in this study will require further analysis, for example, by co-expressing the mutated allele together with the ITPR2 and ITPR3 genes in model systems.
In conclusion, our findings indicate that the c.5360T>C variant causes autosomal recessive and congenital SCA29 with similarities to ataxic cerebral palsy. In a heterozygous state, the variant is associated with cerebellar hypoplasia but does not express clinically. Two heterozygous individuals remained asymptomatic up to the eighth decade. Our observations are in line with previous examples of recessive inheritance in disorders considered to be exclusively dominant,34, 35 and a likely explanation in the family presented herein is a partial loss of IP3R1 channel function caused by the missense variant in heterozygotes. The findings further broaden the correlations between ITPR1 gene variants and their clinical expression as well as subclinical features to be taken into account also in families segregating non-dominant SCA.
References
Durr A : Autosomal dominant cerebellar ataxias: polyglutamine expansions and beyond. Lancet Neurol 2010; 9: 885–894.
Fogel BL, Perlman S : Clinical features and molecular genetics of autosomal recessive cerebellar ataxias. Lancet Neurol 2007; 6: 245–257.
Smeets CJ, Verbeek DS : Climbing fibers in spinocerebellar ataxia: a mechanism for the loss of motor control. Neurobiol Dis 2016; 88: 96–106.
Bezprozvanny I : The inositol 1,4,5-trisphosphate receptors. Cell Calcium 2005; 38: 261–272.
Foskett JK, White C, Cheung KH, Mak DO : Inositol trisphosphate receptor Ca2+ release channels. Physiol Rev 2007; 87: 593–658.
Matsumoto M, Nakagawa T, Inoue T et al: Ataxia and epileptic seizures in mice lacking type 1 inositol 1,4,5-trisphosphate receptor. Nature 1996; 379: 168–171.
Hara K, Shiga A, Nozaki H et al: Total deletion and a missense mutation of ITPR1 in Japanese SCA15 families. Neurology 2008; 71: 547–551.
Iwaki A, Kawano Y, Miura S et al: Heterozygous deletion of ITPR1, but not SUMF1, in spinocerebellar ataxia type 16. J Med Genet 2008; 45: 32–35.
Novak MJ, Sweeney MG, Li A et al: An ITPR1 gene deletion causes spinocerebellar ataxia 15/16: a genetic, clinical and radiological description. Mov Disord 2010; 25: 2176–2182.
Synofzik M, Beetz C, Bauer C et al: Spinocerebellar ataxia type 15: diagnostic assessment, frequency, and phenotypic features. J Med Genet 2011; 48: 407–412.
van de Leemput J, Chandran J, Knight MA et al: Deletion at ITPR1 underlies ataxia in mice and spinocerebellar ataxia 15 in humans. PLoS Genet 2007; 3: e108.
Parolin Schnekenberg R, Perkins EM, Miller JW et al: De novo point mutations in patients diagnosed with ataxic cerebral palsy. Brain 2015; 138: 1817–1832.
Sasaki M, Ohba C, Iai M et al: Sporadic infantile-onset spinocerebellar ataxia caused by missense mutations of the inositol 1,4,5-triphosphate receptor type 1 gene. J Neurol 2015; 262: 1278–1284.
Gerber S, Alzayady KJ, Burglen L et al: Recessive and dominant de novo ITPR1 mutations cause Gillespie syndrome. Am J Hum Genet 2016; 98: 971–980.
Shadrina MI, Shulskaya MV, Klyushnikov SA et al: ITPR1 gene p.Val1553Met mutation in Russian family with mild spinocerebellar ataxia. Cerebellum Ataxias 2016; 3: 2.
McEntagart M, Williamson KA, Rainger JK et al: A restricted repertoire of de novo mutations in ITPR1 cause Gillespie syndrome with evidence for dominant-negative effect. Am J Hum Genet 2016; 98: 981–992.
Tada M, Nishizawa M, Onodera O : Roles of inositol 1,4,5-trisphosphate receptors in spinocerebellar ataxias. Neurochem Int 2016; 94: 1–8.
Huang L, Chardon JW, Carter MT et al: Missense mutations in ITPR1 cause autosomal dominant congenital nonprogressive spinocerebellar ataxia. Orphanet J Rare Dis 2012; 7: 67.
Wang K, Li M, Hakonarson H : ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010; 38: e164.
Adzhubei IA, Schmidt S, Peshkin L et al: A method and server for predicting damaging missense mutations. Nat Methods 2010; 7: 248–249.
Choi Y, Sims GE, Murphy S, Miller JR, Chan AP : Predicting the functional effect of amino acid substitutions and indels. PLoS ONE 2012; 7: e46688.
Schwarz JM, Cooper DN, Schuelke M, Seelow D : MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods 2014; 11: 361–362.
Reese MG, Eeckman FH, Kulp D, Haussler D : Improved splice site detection in Genie. J Comput Biol 1997; 4: 311–323.
Lathrop GM, Lalouel JM, Julier C, Ott J : Strategies for multilocus linkage analysis in humans. Proc Natl Acad Sci USA 1984; 81: 3443–3446.
Pires DE, Ascher DB, Blundell TL : mCSM: predicting the effects of mutations in proteins using graph-based signatures. Bioinformatics 2014; 30: 335–342.
Fan G, Baker ML, Wang Z et al: Gating machinery of InsP3R channels revealed by electron cryomicroscopy. Nature 2015; 527: 336–341.
Lek M, Karczewski K, Minikel E et al: Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016; 536: 285–291.
Lek M, Karczewski KJ, Minikel EV et al: Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016; 536: 285–291.
Ogura H, Matsumoto M, Mikoshiba K : Motor discoordination in mutant mice heterozygous for the type 1 inositol 1,4,5-trisphosphate receptor. Behav Brain Res 2001; 122: 215–219.
Hisatsune C, Miyamoto H, Hirono M et al: IP3R1 deficiency in the cerebellum/brainstem causes basal ganglia-independent dystonia by triggering tonic Purkinje cell firings in mice. Front Neural Circuits 2013; 7: 156.
Sugawara T, Hisatsune C, Le TD et al: Type 1 inositol trisphosphate receptor regulates cerebellar circuits by maintaining the spine morphology of Purkinje cells in adult mice. J Neurosci 2013; 33: 12186–12196.
Furuichi T, Simon-Chazottes D, Fujino I et al: Widespread expression of inositol 1,4,5-trisphosphate receptor type 1 gene (Insp3r1 in the mouse central nervous system. Receptors Channels 1993; 1: 11–24.
Klar J, Hisatsune C, Baig SM et al: Abolished InsP3R2 function inhibits sweat secretion in both humans and mice. J Clin Invest 2014; 124: 4773–4780.
de Vries BB, Pals G, Odink R, Hamel BC : Homozygosity for a FBN1 missense mutation: clinical and molecular evidence for recessive Marfan syndrome. Eur J Hum Genet 2007; 15: 930–935.
Khan TN, Klar J, Tariq M et al: Evidence for autosomal recessive inheritance in SPG3A caused by homozygosity for a novel ATL1 missense mutation. Eur J Hum Genet 2014; 22: 1180–1184.
Acknowledgements
We are particularly grateful to all members of the family who participated in this study and to Dr Anna Grabowska, Department of Radiology, Uppsala University, for reviewing the MRI. We thank the Genome Center platform at Science for Life Laboratory, Uppsala University, for technical support. This work was supported by grants from the Swedish Research Council (2015-02424), Uppsala University Hospital, Science for Life Laboratory. ZA was supported by the Higher Education Commission (HEC) of Pakistan. A video presentation of the study is available as supplementary information on European Journal of Human Genetics website.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Klar, J., Ali, Z., Farooq, M. et al. A missense variant in ITPR1 provides evidence for autosomal recessive SCA29 with asymptomatic cerebellar hypoplasia in carriers. Eur J Hum Genet 25, 848–853 (2017). https://doi.org/10.1038/ejhg.2017.54
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2017.54
This article is cited by
-
The genetic architecture of aniridia and Gillespie syndrome
Human Genetics (2019)