Abstract
Though dysfunction of neuromuscular junction (NMJ) is associated with congenital myasthenic syndrome (CMS), the proteins involved in neuromuscular transmission have not been completely identified. In this study, we aimed to identify a novel CMS gene in a consanguineous family with limb-girdle type CMS. Homozygosity mapping of the novel CMS gene was performed using high-density single-nucleotide polymorphism microarrays. The variants in CMS gene were identified by whole-exome sequencing (WES) and Sanger sequencing. A 20 MB-region of homozygosity (ROH) was mapped on chromosome 6q15–21. This was the only ROH that present in all clinically affected siblings and absent in all clinically unaffected siblings. WES showed a novel variant of AK9 gene located in this ROH. This variant was a start-gain mutation and introduced a cryptic 5′-UTR signal in intron 5 of the AK9 gene. The normal splicing signal would be interfered by the cryptic translation signal leading to defective splicing. Another 25 MB-ROH was found on chromosome 11p13–q12 in all siblings. WES showed a homozygous RAPSN pathogenic variant in this ROH. Since RAPSN-associated limb-girdle type CMS was only manifested in AK9 homozygous variant carriers, the disease phenotype was of digenic inheritance, and was determined by the novel disease modifier AK9 which provides NTPs for N-glycosylation. This is the first time that this specific genotype–phenotype correlation is reported. Importantly, the AK9-associated nucleotide deficiency may replete by dietary supplements. Since AK9 is a disease modifier, enhancing N-glycosylation by increasing dietary nucleotides may be a new therapeutic option for CMS patients.
Similar content being viewed by others
Introduction
Neuromuscular junction (NMJ) of skeletal muscle is a synapse that transmits an impulse from the nerve ending of a motor neuron to the muscle. When an action potential reaches the nerve ending, a neurotransmitter acetylcholine (ACh) is released and ACh will initiate a muscle contraction by binding to nicotinic ACh receptors (AChR) on the muscle membrane. NMJ is clinically important because abnormal neuromuscular transmission can cause myasthenic syndrome. Myasthenic syndrome is characterized by the presence of ocular, bulbar, respiratory symptoms and a decremental response of compound muscle action potential to repetitive nerve stimulation. The phenotype is variable. Some patients can present with mild symptoms while some can be severely affected. The most severe form of myasthenic syndrome can be fatal because of generalized muscle paralysis and respiratory failure. This patient may require intensive care and respiratory support to assist breathing. Myasthenic syndrome can be caused by botulinum toxins, autoantibodies such as anti-AChR antibody, anti-striated muscle antibody, anti-muscle-specific kinase (MuSK) antibody, anti-lipoprotein-related protein 4 (LRP4) antibody and antistriational antibody and single-gene defects collectively called as congenital myasthenic syndrome (CMS).
CMS is characterized by an early onset of the symptoms and the absence of auto-antibodies. CMS is caused by mutations of genes encoding proteins located in the presynaptic, synaptic and postsynaptic regions of NMJ. To date, more than 20 disease genes of CMS have been identified,1 that is, ALG2,2 ALG14,2 AGRN,3 CHAT,4 CHRNA1,5 CHRNB1,5 CHRND,6 CHRNE,7 COLQ,8 DOK7,9 DPAGT1,10 GFPT1,11 LAMB2,12 LRP4,13 MUSK,14 PREPL,15 RAPSN,16 SCN4A,17 SNAP25,18 SYT219 and GMPPB.20 Four of the genes, GFPT1, DPAG1, ALG14 and ALG2 are recently known to cause limb-girdle type CMS by affecting N-glycosylation pathway.21 The product of GFPT1 is N-acetylglucosamine and is the substrate for DPAG1 for the production of UDP-N-acetylglucosamine and the reaction requires UTP. Mutations in ALG2 and ALG14 would impair the mannosylation steps using GDP-mannose as substrates. The synthesis of GDP-mannose requires GTP as reactants.
Though dysfunction of NMJ is clinically significant, the proteins involved in neuromuscular transmission, development, maintenance and assembly of NMJ have not been completely identified. During studies of CMS,22, 23 we have encountered a consanguineous family with CMS in Hong Kong. To identify the novel disease gene, we mapped the disease loci by microarray analysis and identified the variant which affects the function of CMS causing gene by whole-exome sequencing (WES).
Materials and methods
Family
The proband is a 33-year-old male patient who presented with persistent weakness in all limbs since 16 years of age. When first seen, he presented with progressive walking difficulty, easy fatigability and occasionally falls. There was no diurnal variation, diplopia, ptosis, myalgia or bulbar dysfunction. Physical examination showed limb girdle weakness and proximal muscle atrophy. Muscle power was as follows: shoulder: 4/5, elbow: 5-/5, wrist: 5-/5, hip: 5-/5 and knee: 5-/5 symmetrical on both sides. Gower sign was negative. The Tensilon test was positive and the patient was negative for auto-antibodies against nicotinic acetylcholine receptor (AChR). Other autoantibodies associated with myasthenia were not checked. Blood tests showed normal levels of creatinine kinase, lactate and electrolytes. Muscle biopsy was unremarkable. CT thorax showed thymic hyperplasia and therefore video-assisted thoracoscopic surgery thymectomy was performed. Histology of the thymus was unremarkable and although the patient demonstrated clinical improvement in power with pyridostigmine, the muscular atrophy progressed. Taken together, the patient had impaired neuromuscular transmission and myasthenic syndrome. He had three siblings in whom one also shared similar clinical features. In fact, this sibling took his brother's medication by mistake and discovered clinical response to anti-cholinesterase and salbutamol treatment. Since the onset of the disease was in childhood, and there was absence of auto-antibodies, with good response to anticholinesterase treatment, the diagnosis of CMS was established. The clinically affected siblings did not have eye and bulbar signs, limb-girdle type CMS, or else limb-girdle myasthenia was diagnosed. This family had been followed up for 10 years and the clinical phenotypes of all family members remained the same.
Homozygosity mapping for the disease-causing gene
For identifying the region of homozygosity (ROH), high-density single-nucleotide polymorphism (SNP) genotyping array was performed as previously described.24 Whole-genome scan was done with Genome-Wide Human SNP array 6.0 (Affymetrix, Santa Clara, CA, USA) and the data were analyzed with Genotyping Console Software (Affymetrix, Santa Clara, CA, USA) according to the manufacturers’ protocols.
Whole-exome sequencing (WES)
Target enrichment was performed using SureSelectXT Human All Exon V4 target kit (Agilent Technologies) and Nextera kits (Illumina Inc., San Diego, CA, USA) according to the manufacturers’ protocols. Sequencing was performed on an Illumina Inc., HiSeq 2000 sequencer with 100 bp paired end module (Illumina Inc.). WES data analysis was performed using VariantStudio (version 2.2.1, Illumina Inc.). Image analysis and base-calling were performed using the standard Illumina data analysis pipeline Real Time Analysis (RTA) version (RTA) v1.12.4.2. Conversion to fastq file was performed using CASAVA v1.8.2. Reads were mapped to the human reference genome build hg19 (http://genome.ucsc.edu/) using BWA (Burrows-Wheeler Aligner, http://bio-bwa.sourceforge.net/) and SAMtools (http://samtools.sourceforge.net/).
PCR and Sanger sequencing
Genomic DNA of the proband, the parents, and the three siblings were extracted from whole-blood samples by the QIAamp blood kit (Qiagen, Hilden, Germany) according to the manufacturer’s instruction. The study was performed in accordance with the principles of the Declaration of Helsinki. Informed consents were obtained from all subjects. The exon and flanking introns of single-nucleotide variants (SNVs) were PCR amplified. Bi-directional sequencing of purified PCR products was performed by BigDyeDeoxy terminator cycle sequencing reagents according to the manufacturer’s instructions (Life Technologies, Carlsbad, CA, USA). Purified sequencing fragments were separated by capillary electrophoresis and detected via laser-induced fluorescence on an ABI Prism 3100 genetic analyzer (Life Technologies, Carlsbad, CA). The variant was described according to Human Genome Variation Society guidelines on nomenclature for the description of sequence variants (http://www.hgvs.org/mutnomen). Heterozygous indels were decrypted by PrimeIndel.25 The novel SNV was screened in 152 normal ethnic-matched individuals by direct DNA sequencing. All data and phenotype were submitted to Leiden Open Variation Database (LOVD v3.0) - http://databases.lovd.nl/shared/variants/0000097095 and http://databases.lovd.nl/shared/individuals/00078891.
Post-sequencing bioinformatic analysis
WES data filtering was performed using VariantStudio (version 2.2.1, Illumina Inc.). The damaging effects of SNVs on gene products were assessed by PolyPhen-2 and SIFT. N-glycosylation sites were predicted by NetNGlyc. Leiden Open Variation Database 3.0 (LOVD) was used for determining whether the identified variants were reported or not. Tissue expression was searched through BioGPS. Prediction of splice site variant pathogenicity was performed using Human Splicing Finder (HSF version 2.4.1). Promotor and transcription start site were predicted by Neural Network Promoter Prediction (http://www.fruitfly.org/seq_tools/promoter.html). Upstream ORFs were predicted by RegRNA. Scoring of ATG as initiation codons based on Kozak consensus sequence was performed by DNA Functional Sites Miner (DNAFSMiner). Whole-exome sequencing data was extracted from NHLBI Exome Sequencing Project through Exome Variant Server (EVS). Websites of all the software were shown in the Supplementary Table 1. All filtered data of the family members and the proband with haplotype analysis of the AK9 gene are available in the Supplementary Figure 1.
Results
Homozygosity mapping for the disease-causing gene
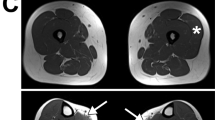
We have performed homozygosity mapping for the disease locus of this consanguineous family. We have mapped the disease gene to a ROH of 20 MB on chromosome 6q15–21 using microarray analysis (Figure 1) which consists of 138 genes. This ROH region is the only ROH region that is present in all the clinically affected siblings and absent in all the clinically unaffected siblings.
Affymetrix SNP 6.0 microarray analysis. The samples of the father (F), mother (M), clinically affected sibling 1 (P1), clinically affected sibling 2 (P2), clinically unaffected sibling 1 (S1), clinically unaffected sibling 2 (S2) were analyzed. Upper panel: The only ROH (region of homozygosity) shared by P1 and P2, colored in green and brown, respectively, was located on chromosome 6q15–21. This is a new CMS locus since no known CMS genes are located in this region: ALG2 on chr. 9q, ALG14 on chr. 1p, AGRN on chr. 1p, CHAT on chr. 10q, CHRNA1 on chr. 2q, CHRNB1 on chr. 17p, CHRND on chr. 2q, CHRNE on chr. 17p, COLQ on chr. 3p, DOK7 on chr. 4p, DPAGT1 on chr. 11q, GFPT1 on chr. 2p, LAMB2 on chr. 3p, LRP4 on chr. 11p, MUSK on chr. 9q, PREPL on chr. 2p, RAPSN on chr. 11p, SCN4A on chr. 17q, SNAP25 on chr. 20p, SYT2 on chr. 1q and GMPPB on chr. 3p. Lower panel: Another ROH region located on chromosome 11p13–q12 was shared among P1, P2, S1 and S2, colored in green, brown, blue, and violet, respectively. Among all known CMS genes, RAPSN gene is located in this region.
Identification of variants which affect protein function
Chromosome 6
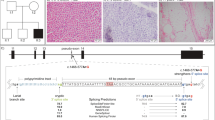
The filtering criteria for recessive pathogenic variants are shown in Figure 2a. A total of 83 variants were located on the ROH region of chromosome 6 and 22 of them passed the family based filtering for autosomal recessive inheritance. After further filtering the variants with minor allele frequency (MAF) <1% in dbSNP Human Build 137 and 1000 Genomes, only five variants remained and two were located in the AK9 gene, a novel SNV NM_001145128.2:c.332-14A>G and a reported SNV c.2191+53C>A (dbSNP ID: rs78959537) and three were indel intronic variants. After bioinformatic interpretation, only the novel SNV NM_001145128.2:c.332-14A>G was predicted to affect protein function. (Figure 2a). The read depth and genotypes of the variant NM_001145128.2:c.332-14A>G in the family is shown in Figure 2b.
Results of ROH on chromosome 6 shared by clinically affected siblings P1 and P2 and ROH on chromosome 11 shared by all siblings (P1, P2, S1, S2). (a) The number of filtered variants remained after each filtering step. (b) WES mutant depth of the filtered SNVs in the family. Sanger sequencing confirmed the WES genotypes of the AK9 (NM_001145128.2) and RAPSN (NM_005055.4, NG_ 008312.1) genes of the family members. The start-gain mutation ATG is underlined in P1 and P2. The parents were doubly heterozygous for AK9 and RAPSN variants. Chr, Chromosome; hom, homozygous; het, heterozygous.
We examined the SNVs in the AK9 gene for haplotype analysis (Supplementary Figure 1). From the constructed haplotypes, S1 is a carrier with the disease chromosome inherited from mother while S2 is a non-carrier. The NM_001145128.2:c.332-14A>G variant and genotypes were confirmed by Sanger sequencing in all family members (Figure 2b). In addition, this variant has not been reported before using exome sequencing data from NHLBI Exome Sequencing Project through the EVS (Supplementary Table 2) and the Exome Aggregation Consortium. However, 25 SNVs of AK9 gene are predicted to be probably damaging with a combined MAF of 0.287% and all of them were only identified in heterozygous states. Assuming Hardy-Weinberg equilibrium, autosomal recessive inheritance, and 100% penetrance, the AK9-associated monogenic disease prevalence is estimated to be 1 in 121 700. This variant was not detected in 152 local Chinese controls using Sanger sequencing (data not shown). According to Clinical Molecular Genetics Society practice guidelines for the interpretation and reporting of unclassified variants (UVs) in clinical molecular genetics, this result indicates the allele frequency of NM_001145128.2:c.332-14A>G is <1% at 95% confidence.
Chromosome 11
Another ROH of 25 MB size was identified on chromosome 11p13–q12 by microarray analysis shared by all siblings (Figure 1). A total of 431 genes were located in this ROH.
We further analyzed the WES data by considering both S1 and S2 to be homozygotes but clinically unaffected. A total of 244 variants were located on the ROH region of chromosome 11 and 40 of them passed the family based filtering for autosomal recessive inheritance. After further filtering the 40 variants with MAF <1% in dbSNP Human Build 137 and 1000 Genomes, only four SNVs and four indel variants remained. The four SNV were RAPSN NM_005055.4:c.491G>A; NP_005046.2:p.(Arg164His), OR4A5 NM_001005272.3:c.220A>G; NP_001005272.3:p.(Thr74Ala), ACCSL NM_001031854.2:c.636-65C>T and CKAP5 NM_001008939.3:c4690-4T>A.
Using PolyPhen-2 prediction, the RAPSN NM_005055.4:c.491G>A; NP_005046.2:p.(Arg164His) was predicted to be probably damaging (score=0.984) and OR4A5 NM_001005272.3:c.220A>G; NP_001005272.3:p.(Thr74Ala) was predicted to be benign (score=0.000). Using SIFT prediction, both SNVs were predicted to be tolerated. The CKAP5 NM_001008939.3:c4690-4T>A and ACCSL NM_001031854.2:c.636-65C>T were predicted to be not damaging using Human Splicing Finder (version 2.4.1). Indel variants were filtered out after bioinformatics interpretation. The RAPSN variant NM_005055.4:c.491G>A and genotypes were confirmed by Sanger sequencing in family members. The read depth and genotyping data in the family are shown in Figure 2b.
Exclusion of X-linked inheritance
Since only male siblings were clinically manifested and not the two female siblings, the inheritance may be X-linked. However, no pathogenic variants were identified on the X-chromosome after data filtering (data not shown).
Translation signal analysis for a cryptic 5′-UTR
The NM_001145128.2(AK9):c.332-14A>G is a novel SNV. The AK9 gene was previously a novel gene C6orf199 with uncharacterized functions. The function of the gene was only recently characterized as AK926 and is involved in maintaining the homeostasis of cellular nucleotides by catalyzing the interconversion of nucleoside phosphates. Since the SNV is located within 10–50 nucleotides upstream of the 3′ acceptor splice site and involved an adenine,27 this SNV may affect splicing. Actually, an A>G variant at position −14 in a branch point sequence of hepatic lipase has been shown to cause defective splicing.28 Other A>G branch point substitutions had been observed in various Mendelian disorders, which caused reduced expression of the normal transcripts.29 However, this A>G transversion does not fall in a typical branch point sequence.
Alternatively, we hypothesize this variant may introduce a cryptic start codon by changing ATA at positions −16 to −14 to ATG, that is, a start-gain mutation. In support of this assertion, a cryptic promoter and transcription start site upstream of the variant at position −329 upstream was predicted by Neural Network Promoter Prediction. The start-gain ATG variant has the highest likelihood score of functionality among all ATG in the cryptic 5′ UTR. In addition, RegRNA identified three upstream ORFs, one of 87 bp from −316 to −230, one of 75 bp from −213 to −139, and the other of 93 bp from −108 to −16 upstream of the variant. An upstream open reading frame (uORF) is a main element of 5′-UTRs.30
Taken together, NM_001145128.2(AK9):c.332-14A>G may create a start codon and introduces a cryptic 5′-UTR of 327 bp within intron 5 (Figure 3). This cryptic 5′-UTR sequence in intron 5 of the AK9 gene probably causes splicing signal to be interfered by translation signal leading to defective splicing of intron 5 or causing transcriptional interference.
The NM_001145128.2(AK9):c.332-14A>G creates a new start codon ATG in intron 5 (shown as ata>G). The promoter sequence was underlined and shown in black and the cryptic transcription start site is shown in red. The uORFs were underlined and shown in red. The cryptic 5′-UTR is of 327 bp in size. This may result in transcriptional interference.
Potential N-glycosylation sites in RAPSN
N-glycosylation prediction tool NetNGlyc predicted several asparagine sites in the RAPSN as candidates for N-glycosylation (Figure 4).
Discussion
Interestingly, when S1 and S2 were assumed to be homozygous carriers similar to P1 and P2, a known missense pathogenic variant in the RAPSN gene was identified. The RAPSN (receptor associated proteins of the synapse, 43 kDa) gene encodes a member of a family of proteins that are receptor associated proteins of the synapse. The function of RAPSN is clustering of AChRs in the NMJ and defective RAPSN causes endplate AChR deficiency.16 Though RAPSN p.(Arg164His) variant has been reported in a patient with CMS (LOVD), p.(Arg164His) variant has never been reported in CMS with predominant limb-girdle pattern or in a homozygous state. A variant at the same codon, p.Arg164Cys, has been shown to diminish AChR co-clustering by 50%.31 The protein damaging effect of p.(Arg164His) is expected to be milder than p.(Arg164Cys) since the wild-type and the mutant residue are both positively charged. Hence, the effect of p.(Arg164His) on AChR co-clustering is probably <50%. This may explain the limb-girdle predominant phenotype.
Since only 2 out of 4 family members with homozygous p.(Arg164His) variant manifested CMS and the other two family members are asymptomatic, that is, the penetrance of RAPSN-associated myasthenia in this family is 50%. Interestingly, only male siblings were clinically manifested and not the two female siblings, this is not compatible with RAPSN-associated CMS in which both male and female patients should be equally affected.32 Hence, the p.(Arg164His) variant is probably a hypomorphic RAPSN allele and a second disease-modifying locus is required for disease penetrance. Similarly, the clinical manifestations of a known RAPSN pathogenic variant, p.N88K are also highly variable, ranged from asymptomatic to severe phenotype. This finding also supports the presence of unknown modifiers for RAPSN-associated CMS.33, 34 In this family, the presence of only ROH on chromosome 6 in the clinically affected homozygous variant carriers and only a novel variant was found in this region strongly suggests AK9 is the disease modifier of RAPSN-associated CMS. Through WES approach, pathogenic and modifier variants had been identified in various diseases conditions, for example, TNNT2 and XIRP2 pathogenic variants in sporadic dilated cardiomyopathy.35 The AK9 variant in this family probably lead to a cryptic 5′-UTR sequence causing splicing signal to be interfered by translation signal leading to defective splicing of intron 5. Indeed, variant of the regulatory elements is recently known to play a significant role in oncogene expression and tumorigenesis.36 Therefore, our novel findings identify a genetic mechanism of RAPSN associated limb-girdle myasthenia. To further consolidate the proposed mechanism, we had retrospectively reviewed genes involved in the NMJ and transmission, for example RYR1 and MTM1, and we did not identify any known variants, which affect protein functions.
Adenylate kinases are a gene family and consisted nine members from AK1 to AK9.37 Three AKs are now known to be associated with disease phenotypes, AK1 (OMIM 103000) causes chronic hemolytic anemia in humans,38 AK2 (OMIM 267500) causes reticular dysgenesis in humans,39 and AK7 (OMIM 615364) with primary ciliary dyskinesia in mice.40 AK1, like AK9, is cytosolic and associated with mental retardation and psychomotor retardation. Taken together, AKs are disease genes and associated with specific phenotypes. To date, no phenomics has been published for AK9 transgenic or knock-out mice. AK9 maintains the homeostasis of nucleotides through interconversion of NDPs and NTPs. We envisage a defective AK9 gene will lead to deficiency in cellular CTP, UTP and GTP resulting in reduced dolichol phosphate production by dolichol kinase as the phosphate donor is CTP,41, 42 reduced production of UDP-N-acetylglucosamine and GDP-mannose, respectively. Taken together, the AK9 variant identified in this family probably cause disease by reducing nucleotide sugars for the N-glycosylation pathway.
Recent studies showed that pathogenic variants in GFPT143 and ALG142 genes result in reduced cell-surface expression of muscle AChR, indicating that N-glycosylation is important for cell-surface protein expression.44 Since RAPSN co-clusters AChRs, RAPSN can be the culprit protein of defective N-glycosylation since the same effect on AChRs will be observed if RAPSN also undergoes N-glycosylation. This is supported by several predicted N-glycosylation sites in RAPSN (Figure 4). Among all the putative N-glycosylation sites, the residue at position 88, that is, p.N88 has the highest score among all the sites. Interestingly, this is also the site where the common RAPSN pathogenic variant p.N88K is located.45 Based on this proposed mechanism, a homozygous RAPSN, p.N88K variant could have the same phenotype, that is, limb-girdle myasthenia, as genetic defects of defective N-glycosylation. Since RAPSN cell-surface expression was not studied in cells with defective N-glycosylation,2, 43 this novel link of AK9, N-glycosylation and RAPSN will pave a new way for studying the biology of NMJ and synapses (Figure 5).
The new link between AK9, N-glycosylation and RAPSN in NMJ. In normal state, AK9 involves in phosphorylation NDP to NTPs. NTPs are substrates for synthesis of NDP-sugars and are the substrates for N-glycosylation of AChR and RAPSN proteins (step 1). A high-surface density of AChR in the NMJ is mediated by RAPSN which is an essential protein for AChR co-clustering (step 2) (upper). A defective AK9 reduces N-glycosylation of AChR and RAPSN proteins while a defective RAPSN impairs the co-clustering of AChR in the post-synaptic membrane (lower). Taken together, this two-hit model explains the myasthenic symptoms developed in P1 and P2. ER, endoplasmic reticulum.
In this family, the homozygous RAPSN and AK9 variants cause CMS with predominant limb-girdle pattern. Since RAPSN-associated limb-girdle type CMS was only manifested in family members with AK9 homozygous variant, the disease phenotype was determined by the novel disease modifier AK9 which provides NTPs for N-glycosylation. Indeed, disease modifier gene can be identified in an unbias way through WES approach, for example recently in atypical Aicardi-Goutieres syndrome.46 In this family, both parents were doubly heterozygous for RAPSN and AK9 variants and were asymptomatic, indicating the digenic inheritance is of autosomal recessive inheritance, this is the first time that this specific genotype-phenotype correlation is reported. A number of heterozygous AK9 probably-damaging missense variants have been identified in normal populations. The AK9 gene should be included in the gene panel for limb-girdle type CMS in addition to genes related to defective N-glycosylation. Indeed, targeted drug treatment on disease modifier in addition to the disease-gene could further enhance the treatment response. Importantly, the AK9-associated nucleotide deficiency may replete by dietary supplements since studies have shown that feeding mice with uridine could increase protein glycosylation.47 By increasing protein glycosylation, we expected the phenotype of P1 and P2 may revert to near normal like the two asymptomatic siblings (ie, S1 and S2). In fact, a similar therapeutic approach had been proposed and tested in congenital disorder of glycosylation with successful outcomes.48 Since AK9 is a disease modifier, enhancing N-glycosylation by increasing dietary nucleotides may improve the symptoms of the patients and may become a new therapeutic option for CMS patients.
References
Cruz PM, Palace J, Beeson D : Congenital myasthenic syndromes and the neuromuscular junction. Curr OpinNeurol 2014; 27: 566–575.
Cossins J, Belaya K, Hicks D et al: Congenital myasthenic syndromes due to mutations in ALG2 and ALG14. Brain: J Neurol 2013; 136: 944–956.
Huze C, Bauche S, Richard P et al: Identification of an agrin mutation that causes congenital myasthenia and affects synapse function. Am J Hum Genet 2009; 85: 155–167.
Ohno K, Tsujino A, Brengman JM et al: Choline acetyltransferase mutations cause myasthenic syndrome associated with episodic apnea in humans. Proc Natl Acad Sci USA 2001; 98: 2017–2022.
Engel AG, Ohno K, Milone M et al: New mutations in acetylcholine receptor subunit genes reveal heterogeneity in the slow-channel congenital myasthenic syndrome. Hum Mol Genet 1996; 5: 1217–1227.
Gomez CM, Maselli RA, Vohra BP et al: Novel delta subunit mutation in slow-channel syndrome causes severe weakness by novel mechanisms. Ann Neurol 2002; 51: 102–112.
Ohno K, Hutchinson DO, Milone M et al: Congenital myasthenic syndrome caused by prolonged acetylcholine receptor channel openings due to a mutation in the M2 domain of the epsilon subunit. Proc Natl Acad Sci USA 1995; 92: 758–762.
Ohno K, Brengman J, Tsujino A, Engel AG : Human endplate acetylcholinesterase deficiency caused by mutations in the collagen-like tail subunit (ColQ) of the asymmetric enzyme. Proc Natl Acad Sci USA 1998; 95: 9654–9659.
Beeson D, Higuchi O, Palace J et al: Dok-7 mutations underlie a neuromuscular junction synaptopathy. Science 2006; 313: 1975–1978.
Belaya K, Finlayson S, Slater CR et al: Mutations in DPAGT1 cause a limb-girdle congenital myasthenic syndrome with tubular aggregates. Am J Hum Genet 2012; 91: 193–201.
Senderek J, Muller JS, Dusl M et al: Hexosamine biosynthetic pathway mutations cause neuromuscular transmission defect. Am J Hum Genet 2011; 88: 162–172.
Maselli RA, Ng JJ, Anderson JA et al: Mutations in LAMB2 causing a severe form of synaptic congenital myasthenic syndrome. J Med Genet 2009; 46: 203–208.
Ohkawara B, Cabrera-Serrano M, Nakata T et al: LRP4 third beta-propeller domain mutations cause novel congenital myasthenia by compromising agrin-mediated MuSK signaling in a position-specific manner. Hum Mol Genet 2014; 23: 1856–1868.
Chevessier F, Faraut B, Ravel-Chapuis A et al: MUSK, a new target for mutations causing congenital myasthenic syndrome. Hum Mol Genet 2004; 13: 3229–3240.
Regal L, Shen XM, Selcen D et al: PREPL deficiency with or without cystinuria causes a novel myasthenic syndrome. Neurology 2014; 82: 1254–1260.
Ohno K, Engel AG, Shen XM et al: Rapsyn mutations in humans cause endplate acetylcholine-receptor deficiency and myasthenic syndrome. Am J Hum Genet 2002; 70: 875–885.
Tsujino A, Maertens C, Ohno K et al: Myasthenic syndrome caused by mutation of the SCN4A sodium channel. Proc Natl Acad Sci USA 2003; 100: 7377–7382.
Shen XM, Selcen D, Brengman J, Engel AG : Mutant SNAP25B causes myasthenia, cortical hyperexcitability, ataxia, and intellectual disability. Neurology 2014.
Herrmann DN, Horvath R, Sowden JE et al: Synaptotagmin 2 mutations cause an autosomal-dominant form of lambert-eaton myasthenic syndrome and nonprogressive motor neuropathy. Am J Hum Genet 2014; 95: 332–339.
Belaya K, Rodríguez PM, Liu WW et al: Mutations in GMPPB cause congenital myasthenic syndrome and bridge myasthenic disorders with dystroglycanopathies. Brain 2015; 138: 2493–2504.
Houlden H : Defective N-linked protein glycosylation pathway in congenital myasthenic syndromes. Brain 2013; 136: 692–695.
Yeung WL, Lam CW, Fung LW, Hon KL, Ng PC : Severe congenital myasthenia gravis of the presynaptic type with choline acetyltransferase mutation in a Chinese infant with respiratory failure. Neonatology 2009; 95: 183–186.
Yeung WL, Lam CW, Ng PC : Intra-familial variation in clinical manifestations and response to ephedrine in siblings with congenital myasthenic syndrome caused by novel COLQ mutations. Dev Med Child Neurol 2010; 52: e243–e244.
Lam CW, Lau KC, Tong SF : Microarrays for personalized genomic medicine. Adv Clin Chem 2010; 52: 1–18.
Lam CW : PrimeIndel: four-prime-number genetic code for indel decryption and sequence read alignment. Clin Chim Acta 2014; 436: 1–4.
Amiri M, Conserva F, Panayiotou C, Karlsson A, Solaroli N : The human adenylate kinase 9 is a nucleoside mono- and diphosphate kinase. Int J Biochem Cell Biol 2013; 45: 925–931.
Harris NL, Senapathy P : Distribution and consensus of branch point signals in eukaryotic genes: a computerized statistical analysis. Nucleic Acids Res 1990; 18: 3015–3019.
Brand K, Dugi KA, Brunzell JD, Nevin DN, Santamarina-Fojo S : A novel A—>G mutation in intron I of the hepatic lipase gene leads to alternative splicing resulting in enzyme deficiency. J Lipid Res 1996; 37: 1213–1223.
Kralovicova J, Lei H, Vorechovsky I : Phenotypic consequences of branch point substitutions. Hum Mutat 2006; 27: 803–813.
Somers J, Poyry T, Willis AE : A perspective on mammalian upstream open reading frame function. Int J Biochem Cell Biol 2013; 45: 1690–1700.
Muller JS, Baumeister SK, Rasic VM et al: Impaired receptor clustering in congenital myasthenic syndrome with novel RAPSN mutations. Neurology 2006; 67: 1159–1164.
Milone M, Shen XM, Selcen D et al: Myasthenic syndrome due to defects in rapsyn: clinical and molecular findings in 39 patients. Neurology 2009; 73: 228–235.
Ku CS, Tan EK, Cooper DN : From the periphery to centre stage: de novo single nucleotide variants play a key role in human genetic disease. J Med Genet 2013; 50: 203–211.
Gallati S : Disease-modifying genes and monogenic disorders: experience in cystic fibrosis. Appl Clin Genet 2014; 7: 133–146.
Long PA, Larsen BT, Evans JM, Olson TM : Exome sequencing identifies pathogenic and modifier mutations in a child with sporadic dilated cardiomyopathy. J Am Heart Assoc 2015; 4: e002443.
Mansour MR, Abraham BJ, Anders L et al: Oncogene regulation. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science 2014; 346: 1373–1377.
Panayiotou C, Solaroli N, Karlsson A : The many isoforms of human adenylate kinases. Int J Biochem Cell Biol 2014; 49: 75–83.
Matsuura S, Igarashi M, Tanizawa Y et al: Human adenylate kinase deficiency associated with hemolytic anemia. A single base substitution affecting solubility and catalytic activity of the cytosolic adenylate kinase. J Biol Chem 1989; 264: 10148–10155.
Lagresle-Peyrou C, Six EM, Picard C et al: Human adenylate kinase 2 deficiency causes a profound hematopoietic defect associated with sensorineural deafness. Nat Genet 2009; 41: 106–111.
Fernandez-Gonzalez A, Kourembanas S, Wyatt TA, Mitsialis SA : Mutation of murine adenylate kinase 7 underlies a primary ciliary dyskinesia phenotype. Am J Respir Cell Mol Biol 2009; 40: 305–313.
Mookerjea S, Coolbear T, Sarkar ML : Key role of dolichol phosphate in glycoprotein biosynthesis. Can J Biochem Cell Biol 1983; 61: 1032–1040.
Shridas P, Waechter CJ : Human dolichol kinase, a polytopic endoplasmic reticulum membrane protein with a cytoplasmically oriented CTP-binding site. J Biol Chem 2006; 281: 31696–31704.
Zoltowska K, Webster R, Finlayson S et al: Mutations in GFPT1 that underlie limb-girdle congenital myasthenic syndrome result in reduced cell-surface expression of muscle AChR. Hum Mol Genet 2013; 22: 2905–2913.
Mellergaard M, Skovbakke SL, Schneider CL et al: N-glycosylation of asparagine 8 regulates surface expression of major histocompatibility complex class I chain-related protein A (MICA) alleles dependent on threonine 24. J Biol Chem 2014; 289: 20078–20091.
Muller JS, Mildner G, Muller-Felber W et al: Rapsyn N88K is a frequent cause of congenital myasthenic syndromes in European patients. Neurology 2003; 60: 1805–1810.
Lessel D, Saha B, Hisama F et al: Atypical Aicardi-Goutieres syndrome: is the WRN locus a modifier? Am J Med Genet Part A 2014; 164A: 2510–2513.
Urasaki Y, Pizzorno G, Le TT : Uridine affects liver protein glycosylation, insulin signaling, and heme biosynthesis. PloS one 2014; 9: e99728.
Freeze HH, Eklund EA, Ng BG et al: Neurology of inherited glycosylation disorders. Lancet Neurol 2012; 11: 453–466.
Acknowledgements
This study was supported by the General Research Fund (GRF), no. 768712 by the Research Grant Countil (RGC) of Hong Kong, China.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on European Journal of Human Genetics website
Rights and permissions
About this article
Cite this article
Lam, CW., Wong, KS., Leung, HW. et al. Limb girdle myasthenia with digenic RAPSN and a novel disease gene AK9 mutations. Eur J Hum Genet 25, 192–199 (2017). https://doi.org/10.1038/ejhg.2016.162
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2016.162
This article is cited by
-
Congenital myasthenic syndromes
Orphanet Journal of Rare Diseases (2019)
-
Adenylate Kinase: A Ubiquitous Enzyme Correlated with Medical Conditions
The Protein Journal (2019)
-
Whole-exome sequencing identifies novel pathogenic mutations and putative phenotype-influencing variants in Polish limb-girdle muscular dystrophy patients
Human Genomics (2018)
-
Myofibrillar myopathy in the genomic context
Journal of Applied Genetics (2018)
-
Mutations in GFPT1-related congenital myasthenic syndromes are associated with synaptic morphological defects and underlie a tubular aggregate myopathy with synaptopathy
Journal of Neurology (2017)