Abstract
Owing to its geographic location near the longitudinal center of Asia, Ladakh, the land of high passes, has witnessed numerous demographic movements during the past millenniums of occupation. In an effort to view Ladakh’s multicultural history from a paternal genetic perspective, we performed a high-resolution Y-chromosomal survey of Ladakh, within the context of Y haplogroup and haplotype distributions of 41 Asian reference populations. The results of this investigation highlight the rich ethnic and genetic diversity of Ladkah which includes genetic contributions from disparate regions of the continent including, West, East, South and Central Asia. The phylogenetic signals from Ladakh are consistent with the Indo-Aryans' occupation during the Neolithic age and its historic connection with Tibet, as well as the East–West gene flow associated with the Silk Road.
Similar content being viewed by others
Introduction
Approximately halfway between the western shores of the Black Sea and the eastern coast of China’s Yellow Sea is Ladakh or ‘the land of high passes’.1 Ladakh is a sub-region of what is now India’s north–west state of Jammu and Kashmir. It is situated on a high plateau, exceeding 3000 meters in most places, and wedged between two mountain ranges, the Kunlun to the north and the Himalayas to the south.1 The narrow Indus River and Hindu Kush corridor cleaves the territory of Ladakh. Although present day Ladakh is one of the most remote areas of the world, it had, for centuries, occupied a strategic location along historical trade routes including the ancient Silk Road. Specifically, the region of Ladakh was the nexus point between Central Asia and South Asia when the Silk Road was utilized. The 60-day journey on the Ladakh route linking Amritsar (Northwest India) and Yarkand (Southwest China) on the southern branch of the silk road was routinely employed by merchants until the late 19 hundreds2 when Mainland China closed the borders. Another common branch of the Silk Road, the Kalimpong route, connected Ladakh with Lhasa, current capital of Tibet, via Gartok, the administrative center of western Tibet. Gartok, in turn, was accessible from the south through the Indus Valley. In addition, the Cherko pass also joined Ladakh to the Lhasa road.
Apart from commercial traffic, this region bore witness to numerous other demographic movements resulting in a rich multicultural history. Genetic results trace the occupation of the Tibetan plateau to the Neolithic age whereas archeological data suggest an earlier habitation in the Late Paleolithic age.3 Mitochodrial DNA markers have confirmed that the vast majority of maternal lineages entered the Tibetan plateau during the final Upper Paleolithic age and early Neolithic age (mid-Holocene) from northern China.3 In addition, the presence of archaic Denisovan-like autosomal elements in Tibetans imply gene flows from northern China.4 Archeological evidence in the form of rock carvings discovered in Ladakh suggest that this area has been occupied since the Neolithic period.1 It is commonly believed that the earliest residents of Ladakh were the Mons of Indian descent and Dards from Iran resulting in a composite Indo-Aryan heritage.1, 5 Before the creation of the kingdom of Ladakh around 950 CE, this territory has been annexed by various political regimes such as the Kushan Empire in the first century AD and Tibet from the mid-seventh to mid-ninth century AD.1, 6 Frequent incursions by the Muslim states of Central Asia during the 14th to 16th centuries1 enhanced the ethnic diversity of the region. Infact Hindu Kush (Hindu slaughter) takes its name from the genocide of Hindus slaves as they were transported to the Muslim courts of Central Asia during the ninth to 18th centuries.
In an effort to chronicle the genetic diversity of Ladakh accumulated from multiple layers of human occupation and dispersals over many millenniums, we present here, for the first time, the high-resolution paternal profile of the Ladkah population. With this genetic snapshot, we hope to alleviate the lacuna of information on Ladakh’s population. For a more thorough interpretation of the data, we also include paternal data from 41 geographically relevant reference populations spanning the Asian continent (Supplementary Table S1).
Materials and methods
Sample collection and DNA extraction
A total of 220 buccal cell swab samples were collected with informed consent from male individuals from Ladakh, India (Supplementary Figure S1). Paternal ancestry for each male donor was recorded for at least two generations. Geographical and linguistic information on the populations is provided in Supplementary Table S1. Genomic DNA was isolated as previously described.7
Y-STR genotyping
The DNA samples were genotyped with a panel of 17 STR loci (DYS19, DYS385a/b, DYS389I/II, DYS390, DYS391, DYS392, DYS393, DYS437, DYS438, DYS439, DYS448, DYS456, DYS458, DYS635 and Y GATA H4) as previously described.8 Amplification reactions, capillary gel electrophoresis, size comparison, analytical designations were performed as indicated in a previous publication.7
Y-SNP genotyping
Eighty-seven binary genetic makers were hierarchically genotyped as previously described.9, 10 The Y-STR and Y-SNP genotype were submitted to the YHRD database under the accession number YA004026. Genetic polymorphisms were assessed as previously described.11 The Y-SNP haplogroup nomenclature follows the recommendation originally proposed by Karafet et al12 and revised with information provided by subsequent scientific teams: Underhill et al13 and the International Society of Genetic Genealogy (www.isogg.org/wiki/Genetic_genealogy). The total number of individuals genotyped for Y-STR markers was 220. Yet, because of the lack of DNA only 209 were genotyped for Y-SNPs.
Statistical and phylogenetic analyses
Thirty seven of the 41 Asian reference populations, listed in Supplementary Table S1, from previously published Y-chromosomal studies were employed to characterize broad phylogeographic patterns using major haplogroups; four populations did not report haplogroup data.
A correspondence analysis (CA) based on the major haplogroups (C/B-R) and a molecular variance analyses were conducted as previously described.11 Frequency contour maps were generated using Surfer software version 12 for the major haplogroups D/H/J/L/O/R (Golden software, Cold Spring Harbor, NY, USA, available from: www.goldensoftware.com). See Supplementary Figure S1 for the location of populations examined in the contour maps.
To view Ladakh’s relative genetic affinities at a finer resolution, pairwise genetic distances (RST values) based on the Y-STR loci (excluding DYS385a/b) were assess. The Ladakh population was compared with a total of 18 geographically pertinent Asian reference populations, listed in Supplementary Table S1, utilizing the Arlequin v3.5 software package (Bern, Switzerland; MyBioSoftware.com).14 This analysis only included 18 populations since to all publications provided Y-STR data. Pairwise population comparisons were performed at a significance level of 0.005 and 1000 permutations15 and Bonferoni correction was applied in order to account for potential type-I errors, (α=0.005/378⩽0.000014). The DYS385 locus and all samples carrying microvariants (STR alleles with a fractional value rather than a whole integer) were excluded from the RST distance calculations. A multidimensional scaling (MDS) plot based on this RST distance matrix was constructed using the XLSTAT software from Addinsoft Corp (www.xlstat.com).
The time estimates (TE) of Y-chromosome haplogroups D-M174, D1a1-N1, D1c-P99, H-M69, H1a*-M82, H*-M69, O-M175, O3a2c1a-M117, R-M207, R1a1a-M198, R1a1a1b2-Z93, R2a-M124, J-M304, L-M20, N-M231, Q-M242 and Q1a2-M346 for thirteen populations were obtained via methods described by Goedbloed et al,16 Sengupta et al17 and Zhivotovsky et al.18 In addition, microsatellite variances were also computed using the intra-population genetic variance (Vp) equation of Kayser et al.19
In order to evaluate genetic relationships between the R1a1a-M198, R2a-M124, D-M174, L-M20 and O3a-P93 individuals, median joining networks were generated at the 15 loci level (DYS385a/b excluded) according to Chennakrishnaiah et al7 and Martinez et al.20
Results
Y haplogroup/haplotype frequencies, diversity and coalescence times
From the 87 total binary genetic makers tested in the 209 Ladakh males, 24 paternal lineages were identified (Figure 1). Of the ten major Y haplogroups (C, D, G, H, J, L, N, O, Q and R) detected in the Ladakh population, the most abundant is the D haplogroup comprising 32% of the individuals. This haplogroup is followed in decreasing order of frequency by R (22%), O (19%) and H (11%). Haplogroups J, N and Q were each found in 4% of the Ladakh males and lower levels of L (2%), C (1%) and G (1%) were also detected. Among the 37 Eurasian populations surveyed for the major Y-chromosomal haplogroups, Ladakh along with Southern Iran (also at 10 haplogroups) is second only to Pakistan (at 12 haplogroups) with respect to the number of haplogroups represented (Supplementary Table S2). Three lineages (D1a1, O3a2c1a, R1a1a1b2 at 23, 17 and 16%, respectively) constitute 56% of the paternal haplotypes. Other polymorphic lineages include D1c and H1a* (9% each) and R2a (5%).
According to the diversity indices (Vp) of the 11 non-overlapping Y-chromosomal haplotypes (Supplementary Table S3) detected in Ladakh, H* exhibited the highest level of variation (Vp=0.9) followed by D1a1-N1 (Vp=0.8), H1a*, L and R2a-M124 (Vp=0.5 each). With a few exceptions including that of H* (40.6±12.4 kya), the relative order of the TE values, in general, follow that of Vp indices. D1a1-N1 (n=58, TE Evo: 42.2±16.9 kya and TE Gen: 16.3±6.5 kya) is estimated to be the oldest haplogroup and, D1c-P99 (n=19, TE Evo: 10.2±2.0 kya and TE Gen: 3.9±0.8 kya), the youngest.
Phylogeography of Y haplogroups/haplotypes
Overall, the principal axis of the CA (Figure 2) displays an east (left) to west (right) orientation. In the far upper-left corner of the plot, Tibet forms a cluster with Northeast, Southeast and South-Asian groups. Near the lower-left center of the graph, there is a cluster encompassing four Central-Asian populations (BUR, MON, KAZ and TUV), a grouping based on the shared polymorphic levels of the C (17–66%) haplogroup. The three populations located in the southwestern-most region of Central Asia (KYR, KAR and SHU) are scattered to the right of the graph just above the center of the y axis. This east–west distribution of populations in the CA parallels that of the L, D, H, O, R and J haplogroup contour gradients (Supplementary Figures S2–S7).
In the MDS plot (Supplementary Figure S8), based on Rst distances (Supplementary Table S4) from Y-STR data, 19 populations representing South (BAN, HAR, J&K, KAT, NEW, LAD, LIN, PUN, VOK and TAM), Central (LHA, QIN and TIB) and West (ARA, GAR, LAV, NAF, SAF and SAS) Asia are represented. Ladakh is located in a tight cluster in the lower-right corner of the plot composed of 11 populations, 6 (BAN, KAT, J&K, LAD, LIN and VOK) from South Asia, 3 (LHA, QIN and TIB) from Central Asia and 2 (GAR and LAV) from West Asia. It is noteworthy that four (HAR, NEW, PUN and TAM) of Ladkah’s geographical South-Asian neighbors lie outside of this cluster and that one of these, Tamang (TAM), an admixed group of Tibetan origin, displays the largest Euclidean distance from Ladakh, of all 19 populations surveyed. The four remaining peripherally located collections (ARA, NAF, SAF and SAS) are from West Asia.
The AMOVA test (Supplementary Table S5) reveals that the highest variation is within populations at 67% (ФST=0.33). However, significant variation also exists among groups (19%, ФCT=0.19), as well as among populations within the groups (14%, ФSC=0.17).
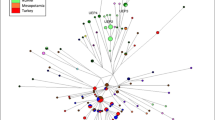
The network phylogenies of R1a1a-M198, L-M20 and R2a-M124 shown in Figures 3, 4 and 5, respectively, feature the haplotypes of Y haplogroups common to the more western regions of our reference area. The network of R2a-M124 (Figure 5), found in polymorphic frequencies across South Asia, Central Asia and West Asia displays a quasi-star like appearance radiating from a degenerate center (no samples present at the central node). There is little population or regional clustering except for a Sasun assembly occupying a terminal position on one of the four major branches. The R2a-M124 network is characterized by a nearly complete absence of inter-population (except for one lineage detected in both collections from Afghanistan) and no inter-regional haplotype sharing. The majority of the nodes are singletons from South, Central and West Asia in that order. South Asian lineages occupy nearly all of the nodes closest to the network origin. However, the Ladakh haplotypes are terminal to those from either South- or Central-Asian groups. The R2a-M124 lineages from the four West-Asian groups (LAV, NAF, SAF and SAS) are also terminal and are not internal to any of the South- or Central-Asian haplotypes.
The R1a1a-M198 network (Figure 3), composed of lineages from South (KAT, LAD, LING, NEW, TAM and VOK), Central (TIB) and three West-Asian populations (NAF, SAF and ARA) are grouped in a dense assembly with no definite center and little terminal clustering. There is a much higher level of haplotype sharing compared with the R2a-M124, both within and among populations of the same regions but none were detected among different geographical groups. Nevertheless, numerous singletons representing all of the populations are scattered throughout the network. As in the R2a-M124 phylogeny, the Ladakh haplotypes occupy mostly terminal positions. However, in contrast to the R2a-M124 network, the haplotypes of South and North Afghanistan (West Asia) are internal to South, Central and other West-Asian nodes. Also, most terminal West-Asian haplotypes germinate from other West-Asian collections. This is not surprising considering that the R1a1a-M198 mutation arose in either East Europe8, 21, 22 or, alternately, a southern region of Central Asia13, 23 and is, today, very common in West Asia. As expected, and as we recently reported for Central and South-Asian individuals,23 the majority of the R1a1a-M198 Ladakh individuals (100% or 35/35) were R-Z93 (the Asian subclade). The R-Z283 European subclade is missing in Ladakh.
The network based on individuals under haplogroup L-M20 (Figure 4) exhibits a bipartite topology. All individuals in the upper cluster associate with the M357 mutation whereas the lower cluster with M76 mutation. In the upper cluster, we identified most of the individuals from Ladakh along with the neighboring populations of SAF and NAF across the Hindu Kush corridor to the west. In addition, people from the Southwest-Indian population of VOK are seen. In the lower cluster, the populations from further west in Armenia (ARA, GAR, LAV and SAS), as well as all the samples from the Lingayat population and most of the Vokkaliga individuals of Southwest India are observed. All of the individuals from Ladakh radiate from West- or South-Asian haplotypes. This partitioning of L-M20 into two distinct conglomerates parallels the distribution of this haplogroup in the corresponding contour map (Supplementary Figure S2) that exhibits frequency foci in Southwest India and West Asia. Little haplotype sharing and partition within the two assemblages is seen in L-M20.
The O3a-P93 network (Supplementary Figure S9) is made up of Sino-Tibetan (NEW, TAM and TIB), Indo-European from Nepal (KAT) and Ladakh. This network presents limited haplotype sharing and no partitioning of populations.
The network based on D-M174 individuals is illustrated in Supplementary Figure S10 and is entirely compose of individuals from Tibet and Ladakh. This projection exhibits a tripartite topology with individuals from both populations in all three clusters. All three assemblages exhibit quasi-star topologies with some Tibetan individuals deriving from Ladakh haplotypes and vice versa. The bottom left and middle clusters are made up of D1a-M15 people whereas the top right conglomerate is composed of D1c-P99 samples. It is interesting that haplotype sharing is observed only among D1a-M15 individuals. These results are consistent with extensive gene flow between Tibet and Ladakh along the East–West road. These data also corroborate the frequency distribution observed in the corresponding contour map (Supplementary Figure S3).
Discussion
Ladakh in northern India, a region bisected by the narrow Indus River Valley and the Hindu Kush corridor, was found to exhibit elevated levels of Y-chromosome diversity. Of all the populations examined in this study (see Supplementary Table S1 for articles24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38 citing the reference populations), Ladakh, Southern Iran and Pakistan, exhibit the highest Y-chromosome diversity. Interesting, these three populations lie in a region of genetic confluence geographically located in west–central Asia. Also noteworthy is that the four major polymorphic Y-chromosomal haplogroups detected in Ladkakh (O=19%, D=32%, R=22% and H=11%) are each representative of a different geographical region of Asia (East, Central, West and South, respectively). These distribution patterns are also reflected in the corresponding Y haplogroup contour gradient maps (Supplementary Figures S5, S3, S6 and S4, respectively). Our data indicate that the paternal ancestry of Ladakh is a genetically diverse mosaic laid down in multiple time intervals from different sources. This inference is also supported by the results of the CA (Figure 2), a graphical representation of genetic relationships. In this analysis, Ladakh fails to aggregate with other populations in the graph and partitions at an intermediate position, proximal to the horizontal center of the CA plot between the Northeast/Southeast Asian conglomerate (populations clustering at the top-left quadrant) and the Near East/West-Asian/South-Asian cluster (populations segregating at the top right quadrant). This position may reflect Ladakh’s composite of East, South and West-Asian Y haplogroups.
The MDS plot (Supplementary Figure S8) and the network phylogenies (Figures 3, 4, 5, Supplementary Figures S9 and S10) afford a closer view of relative genetic affinities between Ladakh and the other Asian populations. In the MDS graph, Ladakh is located near the center of a tight West/South/Central-Asian cluster of eleven groups (BAN, GAR, KAT, J&K, LAD, LAV, LHA, LIN, QIN, TIB and VOK) which suggests extensive inter-population gene flow among these regions. Notable is the close Euclidean positions of Ladakh and Tibet which is also supported by the topology and the co-segregation of Tibet and Ladakh in the D-M174 network. The proximity of these two populations on the MDS plot and the partitioning in the D-M174 network is not surprising given that over the last millennium their history has been closely intertwined and that Ladakh owes a large portion of its genetic heritage to Tibetan colonists especially from the middle of last century. However, groups from other regions of Asia have also contributed to the current Y-chromosomal constitution of the region, an observation reflected by the Ladakh’s proximity to both West- (GAR, LAV) and South-Asian populations (LIN, VOK, J&K, KAT). Further, although it is difficult to pinpoint the absolute time intervals, Ladakh’s more terminally placed lineages relative to the internal Middle Eastern nodes of the R-M198 network (Figure 3) suggest a primarily west to east movement of this haplotype. These graphical results may also reflect the initial colonizations by Mons of Indian descent and Dards from Iran1 and the commercial traffic generated by the Silk Road. In this regard, the rare R-M335 mutation in Ladakh is of interest as a few individuals distributed on an East to West axis (more leading to the East) in Europe, India, Near East, Central Asia and Southeast Asia have been observed and as such may represent a signal from Silk Road gene flow. Together these data portray present day Ladakh as a rich and varied Y-chromosomal landscape, which is, likely, the result of a multi-layered accumulation of demographic episodes (nomadic migration, seasonal pastoral movements, military campaigns, refugee immigrations and trade) each leaving its genetic mark during the millenniums of occupation. Similar observations were made by Di Cristofaro et al in their study of populations from the Afghan Hindu Kush region.39 All together, the lack of partitioning, the small number of mutational steps connecting haplotypes and the limited level of haplotype sharing among populations within the networks from all investigated haplogroups suggest recent gene flow.
References
Jina PS : Ladakh: The Land and the People. Ladakh, India: Indus Publishing, 1996.
Rizvi J : Ladakh, Trans-Himalayan Caravans-Merchant Princes and Peasant Traders in Ladakh. Oxford, England: Oxford University Press, 2001.
Zhaoa M, Qing-Peng K, Hua-Wei W et al: Mitochondrial genome evidence reveals successful Late Paleolithic settlement on the Tibetan Plateau. Proc Natl Acad Sci USA 2009; 106: 21230–21235.
Huerta-Sánchez E, Xin J, Zhuoma AB et al: Altitude adaptation in Tibetans caused by introgression of Denisovan-like DNA, 2014 Nature 512: 194–197.
Kuz’mina EE Mallory JP, Koninklijke Brill NV The Origins of the Indo-Iranians. Leiden, The Netherlands: Hotei Publishing, 2007; Volume 3.
Rizvi J : Ladakh, Crossroads of High Asia. Oxford, England: Oxford University Press, 1996.
Chennakrishnaiah S, Perez D, Gayden T, Rivera L, Regueiro M, Herrera RJ : Indigenous and foreign Y-chromosomes characterize the Lingayat and Vokkaliga populations of Southwest India. Gene 2013; 526: 96–106.
Lacau H, Gayden T, Regueiro M et al: Afghanistan from a Y-chromosome perspective. Eur J Hum Genet 2012; 20: 1063–1070.
Martinez L, Reategui EP, Fonseca LR et al: Superimposing polymorphism: the case of a point mutation within a polymorphic Alu insertion. Hum Hered 2005; 59: 109–117.
Hammer MF, Horai S : Y chromosomal DNA variation and the peopling of Japan. Am J Hum Genet 1995; 56: 951–962.
Gayden T, Cadenas AM, Regueiro M et al: The Himalayas as a directional barrier to gene flow. Am J Hum Genet 2007; 80: 884–894.
Karafet TM, Mendez FL, Meilerman MB, Underhill PA, Zegura SL, Hammer MF : New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree. Genome Res 2008; 18: 830–838.
Underhill PA, Myres NM, Rootsi S et al: A major Y-chromosome haplogroup R1b Holocene era founder effect in Central and Western Europe. Eur J Hum Genet 2011; 19: 95–101.
Excoffier L, Lischer H : Arlequin suite v 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 2010; 10: 564–567.
Kayser M, Brauer S, Stoneking M : A genome scan to detect candidate regions influenced by local natural selection in human populations. Mol Biol Evol 2003; 20: 893–900.
Goedbloed M, Vermeulen M, Fang RN et al: Comprehensive mutation analysis of 17 Y-chromosomal short tandem repeat polymorphisms included in the AmpFISTR Yfiler PCR amplification kit. Int J Legal Med 2009; 123: 471–482.
Sengupta S, Zhivotovsky LA, King R et al: Polarity and temporality of high-resolution y-chromosome distributions in India identify both indigenous and exogenous expansions and reveal minor genetic influence of Central Asian pastoralists. Am J Hum Genet 2006; 78: 202–221.
Zhivotovsky LA, Underhill PA, Cinnioğlu C et al: The effective mutation rate at Y chromosome short tandem repeats, with application to human population-divergence time. Am J Hum Genet 2004; 74: 50–61.
Kayser M, Krawczak M, Excoffier L et al: An extensive analysis of Y-chromosomal microsatellite haplotypes in globally dispersed human populations. Am J Hum Genet 2001; 68: 990–1018.
Martinez L, Underhill PA, Zhivotovsky LA et al: Paleolithic Y-haplogroup heritage predominates in a Cretan highland plateau. Eur J Hum Genet 2007; 15: 485–493.
Semino O, Passarino G, Oefner PJ et al: The genetic legacy of Paleolithic Homo sapiens sapiens in extant Europeans: a Y chromosome perspective. Science 2000; 290: 1155–1159.
Mirabal S, Regueiro M, Cadenas AM et al: Y-chromosome distribution within the geo-linguistic landscape of northwestern Russia. Eur J Hum Genet 2009; 17: 1260–1273.
Underhill PM, Poznik GD, Rootsi S et al: The phylogenetic and geographic structure of Y-chromosome haplogroup R1a. Eur J Hum Genet 2015; 23: 124–131.
Regueiro M, Cadenas AM, Gayden T, Underhill PA, Herrera RJ : Iran: tricontinental nexus for Y-chromosome driven migration. Hum Hered 2006; 61: 132–143.
Al-Zahery N, Semino O, Benuzzi G, Magri C, Passarino G, Torroni A : Santachiara-Ben erecetti AS:Y-chromosome and mtDNA polymorphisms in Iraq, a crossroad of the early human dispersal and of post-Neolithic migrations. Mol Phylogenet Evol 2003; 28: 458–472.
Wells RS, Yuldasheva N, Ruzibakiev R et al: The Eurasian Heartland: a continental perspective on Y-chromosome diversity. Proc Natl Acad Sci USA 2001; 98: 10244–10249.
Karafet T, Xu L, Du R et al: Paternal population history of East Asia: sources, patterns, and microevolutionary processes. Am J Hum Genet 2001; 69: 615–628.
Zhang Y-B, Li X, Zhang F, Wang D-M, Yu J : A preliminary study of copy number variation in Tibetans. PLoS ONE 2012; 7: e41768.
Zhu Y, Lu S, Xie Z, Chen Y, You J : Genetic analysis of 15 STR loci in the population of Zhejiang Province (Southeast China). Forensic Sci Int Genet 2009; 3: 139–140.
Cordaux R, Weiss G, Saha N, Stoneking M : The northeast Indian passageway: a barrier or corridor for human migrations? Mol Biol Evol 2004; 21: 1525–1533.
Kivisild T, Rootsi S, Metspalu M et al: The genetic heritage of the earliest settlers persists both in Indian tribal and caste populations. Am J Hum Genet 2003; 72: 313–332.
Yadav B, Raina A, Dogra TD : Genetic polymorphisms for 17 Y-chromosomal STR haplotypes in Jammu and Kashmir Saraswat Brahmin population. Leg Med 2010; 12: 249–255.
Nagy M, Henke L, Henke J et al: Searching for the origin of Romanies: Slovakian Romani, jats of haryana and jat sikhs Y-STR data in comparison with different Romani populations. Forensic Sci Int 2006; 169: 19–26.
Karafet TM, Meilerman MB, Underhill PA, Zegura SL, Hammer MF : New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree. Genome Res 2008; 18: 830–838.
Hammer MF, Karafet TM, Park H et al: Dual origins of the Japanese: common ground for hunter-gatherer and farmer Y chromosomes. J Hum Genet 2006; 51: 47–58.
Xue Y, Zerjal T, Bao W et al: Male demography in East Asia: a north-south contrast in human population expansion times. Genetics 2006; 172: 2431–2439.
Karafet TM, Lansing JS, Redd AJ : Balinese Y-chromosome perspective on the peopling of Indonesia: genetic contributions from pre-neolithic hunter-gatherers, Austronesian farmers, and Indian traders. Hum Biol 2005; 77: 93–114.
Lowery RK, Herrera K, Uribe G, Reguiero M, Herrera RJ : Sub-population structure evident in forensic Y-STR profiles from Armenian geographical groups. Legal Med 2013; 15: 85–90.
Di Cristofaro J, Pennarun E, Mazières S, Myres NM, Lin AA : Afghan Hindu Kush: where Eurasian sub-continent gene flows converge. PLoS One 2013; 8: e76748.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Web Sources
www.isogg.org/wiki/Genetic_genealogy
Supplementary Information accompanies this paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Rowold, D., Benedico, D., Garcia-Bertrand, R. et al. Ladakh, India: the land of high passes and genetic heterogeneity reveals a confluence of migrations. Eur J Hum Genet 24, 442–449 (2016). https://doi.org/10.1038/ejhg.2015.80
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2015.80
This article is cited by
-
High-altitude population neonatal and maternal phenotypes associated with birthweight protection
Pediatric Research (2022)
-
The genetic affinities of Gujjar and Ladakhi populations of India
Scientific Reports (2020)