Abstract
Foxg1 gene encodes for a transcription factor essential for telencephalon development in the embryonic mammalian forebrain. Its complete absence is embryonic lethal while Foxg1 heterozygous mice are viable but display microcephaly, altered hippocampal neurogenesis and behavioral and cognitive deficiencies. In order to evaluate the effects of Foxg1 alteration in adult brain, we performed expression profiling in total brains from Foxg1+/− heterozygous mutants and wild-type littermates. We identified statistically significant differences in expression levels for 466 transcripts (P<0.001), 29 of which showed a fold change ≥1.5. Among the differentially expressed genes was found a group of genes expressed in the basal ganglia and involved in the control of movements. A relevant (three to sevenfold changes) and statistically significant increase of expression, confirmed by qRT-PCR, was found in two highly correlated genes with expression restricted to the hypothalamus: Oxytocin (Oxt) and Arginine vasopressin (Avp). These neuropeptides have an important role in maternal and social behavior, and their alteration is associated with impaired social interaction and autistic behavior. In addition, Neuronatin (Nnat) levels appear significantly higher both in Foxg1+/− whole brain and in hippocampal neurons after silencing Foxg1, strongly suggesting that it is directly or indirectly repressed by Foxg1. During fetal and neonatal brain development, Nnat may regulate neuronal excitability, receptor trafficking and calcium-dependent signaling and, in the adult brain, it is predominantly expressed in parvalbumin-positive GABAergic interneurons. Overall, these results implicate the overexpression of a group of neuropeptides in the basal ganglia, hypothalamus, cortex and hippocampus in the pathogenesis FOXG1 behavioral impairments.
Similar content being viewed by others
Introduction
Rett syndrome (RTT, OMIM#312750) is a postnatal neurodevelopmental disorder that affects normal brain development during early childhood with an incidence of 1 in 10 000 live female births and represents one of the most common causes of mental retardation in girls.1 This syndrome is characterized by high clinical variability revealing a wide spectrum of clinical variants beside the classical form: the early-onset seizure variant, the Zappella variant (Z-RTT), the congenital variant, the late regression variant and the ‘forme fruste’ variant.2, 3, 4, 5, 6 In 2008, Ariani et al.7 demonstrated that mutations in FOXG1 (forkhead box protein G1) gene are responsible for the congenital variant of RTT. Together with the early-onset seizures variant, the congenital form is the most severe variant.1, 8
The FOXG1 gene encodes for a DNA-binding transcription factor essential for the development of the telencephalon in the embryonic mammalian forebrain that directly regulates gene transcription as well as chromatin dynamics.9 It is one of the first transcription factors activated during telencephalic development, acting as an integrator of signaling centers.10, 11, 12 Its role has been extensively characterized in mice demonstrating that it is implicated in regulating cortical arealization, expansion of the cortical progenitor pool and regulation of progenitor cell cycle length.13, 14, 15, 16, 17, 18 In particular, the protein acts by maintaining telencephalic progenitor status and ensuring that these progenitors maintain appropriate cell cycle kinetics.17 As a consequence of these essential functions in telencephalic development, brains from Foxg1 KO mice show a significant reduction in size of the telencephalic vesicles because of a severely compromised telencephalon growth.11, 19 Despite its early expression in telencephalon development, several investigators demonstrated that FOXG1 expression is detectable in the differentiating cortical compartment and in the hippocampus in the postnatal stages, although at lower levels with respect to the early embryonic phases, suggesting that it may have additional functions in differentiating and mature neurons and in global chromatin organization.7, 20, 21
Despite the important advances in understanding the functions of FoxG1, very little is known about its target genes and only a few of them have been identified to date. In order to identify new targets and possibly the pathways altered by mutations in Foxg1, we thus performed genome-wide transcriptional analysis on RNA from the brain of heterozygous mutant Foxg1 mice.13 Moreover, considering the specific role of Foxg1 in the hippocampus during postnatal neurogenesis, we confirmed our findings in primary hippocampal neurons in which Foxg1 had been acutely silenced by shRNA.
Materials and methods
Mouse models
Foxg1-Cre transgenic animals were used where the Cre recombinase is knocked in the Foxg1 locus replacing the entire Foxg1 intron-less coding sequence.22 Brains from three heterozygous Foxg1-Cre mutant mice and three wild-type littermates at P30 were isolated at Ospedale San Raffaele Scientific Institute, Milan, Italy.22 All experiments were approved by the San Raffaele Scientific Institute Animal Care and Use Committee (IACUC) with Permit Number 603. The experimental protocols adhered to the national and international laws and provisions regarding the protection of animals and all animal experiments were performed by authorized personnel under the rules of Italy according to EC Directive 86/609.
Primary hippocampal neurons
Neuronal cultures were prepared from the hippocampus of day 18.5 mouse embryos (E18.5). Approximately 7 × 104 cells were plated on coverslips coated with poly-L-lysine in 12-well plates and cultured in Neurobasal medium (Invitrogen, Carlsbad, CA, USA) supplemented with B27 (Invitrogen) and glutamine (Sigma-Aldrich, St Louis, MO, USA). The lentiviral knockdown constructs were generated by cloning oligo duplexes into the pLentiLox 3.7 (pLL 3.7) plasmid (http://www.sciencegateway.org/protocols/lentivirus/cloning.htm). The sh-FoxG1#1 construct targets the region 5′-AGGATAGGCCTGGAGTTGTA-3′, the sh-FoxG1#2 construct is directed against the target region 5′-TGATAGCACAGTTGGTACA-3′ and the sh-control construct is directed against Renilla luciferase and targets the region 5′-GGCCTTTCACTACTCCTAC-3′. Primary hippocampal neurons were infected between day 3–6 and analyzed at 14 and 21 days after plating. A transfection with the control shRNA was carried out simultaneously on the same batch of primary neurons. Foxg1 mRNA and protein levels following shRNA treatment were evaluated by qRT-PCR and western blot, respectively, in order to assess Foxg1 silencing (Supplementary Figure 1).
RNA isolation
Total RNA was extracted from the brain or cell cultures with RNeasy mini kit according to the manufacturer’s protocol (Qiagen, Valencia, CA, USA) and quantified using Nanodrop Spectrophotometer ND-1000 (NanoDrop products, Wilmington, DE, USA). Integrity of the total RNA was evaluated using the RNA 6000 Nano Assay Kit on a 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA).
Microarray analysis
Total RNA from the mouse brain was reverse-transcribed, labeled with biotin and amplified overnight using the Illumina TotalPrep RNA Amplification Kit (Ambion, Austin, TX, USA) according to the manufacturer’s protocol. A mixture of 1.5 ug of the biotinylated cRNA sample and Hyb E1 hybridization buffer containing 37.5% (w/w) formamide was hybridized on a mouse WG6 chip on an Illumina platform (Illumina, San Diego, CA, USA) as described.23 Three biological and two technical replicates were performed. BRB array tool was used to identify differentially expressed genes with a detection P value <0.001. Selection of genes for subsequent validation was performed on the basis of the fold change value (>1.3 or <0.7) and of biological function (genes expressed in brain and involved in neurobiological processes or genes already associated to Rett or other intellectual disabilities). Gene Ontology analysis was used to identify genes related to common pathways.
Quantitative polymerase chain reaction (qPCR)
Ten micrograms of total RNA were reverse-transcribed with high capacity cDNA reverse transcription kit according to manufacturer’s instructions (Applied Biosystem, Foster City, CA, USA). Real-time PCR analysis was performed using commercial TaqMan probes (Applied Biosystem, Supplementary Table 1). PCR was carried out in single-plex reactions in a 96-well optical plate with TaqMan Universal PCR Master Mix (Applied Biosystem) on ABI Prism 7700 Sequence Detection System (Applied Biosystems). Experiments were performed in triplicate in a final volume of 20 μl with 25 ng of cDNA template. Standard thermal cycling conditions were used (Applied Biosytems): 2 min at 50 °C and 10 min at 95 °C followed by 40 cycles at 95 °C for 15 s and 60 °C for 1 min. Gene expression values were normalized to expression of Glucuronidase β (GUSB, assay id: Mm00446956_m1). Results were analyzed using the comparative Ct method. The Student’s t-test with a significance level of 95% was used for the identification of statistically significant differences in expression levels among samples.
Immunoblotting
Cells were lysed in RIPA buffer (1% NP-40, 0,5% Na-deoxycholate, 0,1% SDS, 2 mM EDTA, 150 mM NaCl, 50 mM NaF, 50 mM Tris-HCl, pH 7.4, and protease inhibitors). Lysates were centrifuged (200 000 g for 20 min) and quantified and then used for SDS-PAGE electrophoresis. Proteins were separated by SDS-PAGE on 8% acrylamide gels and then transferred to nitrocellulose membranes using standard procedures. The membrane was hybridized overnight with the anti-FOXG1 antibody (ab18259, Abcam, Cambridge, UK) 1:1000 in 1 × TBS/0,05% Tween-20/1% BSA, and then for 1 h with the secondary antibody (1:1000 in 1 × TBS/0,05% Tween-20/1% BSA) conjugated with horseradish peroxidase (GE Healthcare, Little Chalfont Buckinghamshire, England), followed by chemiluminescent revelation. Band quantification was performed with Photoshop. Band intensities were normalized to β-actin signal to correct for differences in the amount of loaded protein.
Immunohistochemistry
Mice were intraperitoneally injected with 200 mg/kg avertin and transcardially perfused with 0.9% saline, followed by ice-cold 4% PFA (Sigma-Aldrich). Brains were post-fixed overnight in 4% PFA and cryoprotected in 30% sucrose (Sigma-Aldrich) at 4 °C for 2 days. Forty-micrometer coronal sections were quenched for endogenous peroxidase activity with 3% H2O2 for 30 min at room temperature, washed in PBS, incubated in blocking solution (10% goat serum, 0.1% Triton in PBS) for 1 h and incubated overnight with primary antibodies (1:100 dilution) at 4 °C. Anti-Oxytocin (ab2078, Abcam), anti-Neuronatin (ab2078, Abcam) and anti-Arginine vasopressin (AB1565, EMD Millipore Corporation, Temecula, CA, USA) were used as primary antibodies. The following day, sections were incubated with Goat anti-Rabbit IgG (H+L) Secondary Antibody, HRP conjugate (Thermo Scientific, Waltham, MA, USA) for 1 h and stained with the VECTASTAIN Elite ABC system kit (Vector Labs, Burlingame, CA, USA).
Statistical analysis
Comparison of expression profiles from Foxg1+/− heterozygous mice and Foxg1+/+ littermates was performed using BRB-ArrayTools Version 3.8.1 (http://linus.nci.nih.gov/BRB-ArrayTools.html). Differences in mRNA levels determined in qPCR were assessed by Student’s t-test with a significance level of 95%. Correlation between microarray and qPCR results was assessed using Pearson’s coefficient.
Results
Microarray analysis of total RNA extracted from the whole brain of three Foxg1+/− heterozygous mutants compared with three wild-type littermates at postnatal day 30 revealed statistically significant differences in expression levels for 466 transcripts (P<0.001, Supplementary Table 2).
Among the 466 differentially expressed genes, 29 showed a fold change ≥1.5 for upregulated genes and <0.5 for downregulated genes. Sixty-three genes showed borderline values of fold change (from 1.3 to 1.5 or from 0.7 to 0.5) (Figure 1). Among them, Mef2c (myocyte enhancer factor 2C) showed a trend toward downregulation that did not reach the defined Fold Change threshold; it was, however, included in the validation set as it has been found mutated in patients with a RTT-like phenotype.24, 25
Among these 92 genes, a set of 11 genes was selected based on their biological function for further validation by qRT-PCR (Table 1) in the same brain samples. qPCR analyses showed a high correlation between the results of the two experiments (r=0.948, P=2.035 × 10−6; Pearson’s correlation test).
A statistically significant alteration of expression between Foxg1+/− mice and wild-type littermates was confirmed for three genes: Oxytocin (Oxt, 9.08±0.45 in Foxg1+/− vs 1.15±0.06 in wild type), Arginine vasopressin (Avp, 2.21±0.03 in Foxg1+/− vs 1.13±0.06 in wild type) and Neuronatin (Nnat, 1.91±0.03 in Foxg1+/− vs 0.91±0.06 in wild type) (Figure 2a). Mef2c reduction in expression was again borderline, owing to considerable variability in expression among the two brain samples that were available for analysis (0.47±0.05 in the first Foxg1+/− vs 1.00±0.02 in wild type and 0.86±0.05 in the second Foxg1+/− vs 1.00±0.18 in wild type, Figure 2b). The expression of Oxt, Avp and Nnat was also evaluated at the protein level and the results for the three genes recapitulate the qPCR data (Figure 3).
(a) Relative quantification of mRNA levels of OXT, AVP, NNAT and MEF2C genes assessed by qRT-PCR using commercial Taqman probes. Analyses were performed in triplicate on the same tissues used for cDNA microarray experiments and data are reported as mean±SEM, *P<0.01; **P<0.05. (b) Relative quantification of Mef2c mRNA levels by qRT-PCR in two out of three samples (mean ± SD, *P<0,005). RNA from one of the foxg1+/− samples was not sufficient for analyses and the validation was thus performed on two samples only.
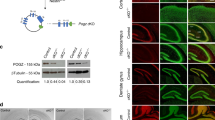
(a–f) Neuronatin (NNAT), Oxytocin (OXT) and Vasopressin (ASP) immunoreactivity in ventromedial hypothalami (VMH) of Foxg1+/+ and Foxg1+/− transgenic mice. (g) Schematic representation depicting the VMH (red square) in a brain coronal section. Scale bar=100 μm. A full color version of this figure is available at the European Journal of Human Genetics journal online.
Given the difficulty in obtaining postmortem brain tissue from affected girls and in distinguishing in those tissues between alterations that are primary causes of disease and those that are secondary effects due to disease progression, we decided to validate the results obtained with microarray analysis in primary hippocampal neurons in which Foxg1 had been silenced by shRNA technology. As Oxt and Avp genes are expressed exclusively in the hypothalamus, qPCR in Foxg1-silenced hippocampal neurons was performed only for Nnat and Mef2c. Interestingly, Nnat that was overexpressed in Foxg1 +/− brains was also upregulated in silenced neurons (Figure 4). A trend toward expression alteration was again observed for Mef2c but the change went in the opposite direction compared with whole brain (reduced expression in whole brain vs increased expression in neurons) (Figure 4).
Discussion
FOXG1 is a transcriptional factor involved in the early forebrain development, where it promotes progenitor proliferation and suppresses premature neurogenesis. FOXG1 function in developing brain has been extensively characterized. However, little is known about its function in postnatal brain, although a consistent expression in adult dentate gyrus and in the subventricular zone of the hippocampus has been reported. In these regions, Foxg1 seems to regulate postnatal neurogenesis.20 In order to characterize the consequences of Foxg1 absence in postnatal brain, herein, we performed global expression profiling in the brain of Foxg1+/− heterozygous mice and Foxg1+/+ littermates at postnatal day 30.
Our expression profiling experiments revealed a set of genes that could be considered direct or indirect Foxg1 interactors. A significant alteration of Oxt and Avp genes was observed in Foxg1+/− brains. These neuropeptides, represented in bold in Table 1, show a very restricted expression pattern that is limited to the hypothalamus 26 and have an important role in maternal and social behavior.27 Alteration of Oxytocin-Vasopressin levels is indeed associated to impaired social interaction and a role for these genes in autistic behavior has been reported.26, 28 The presence of autistic features, like poor eye contact, is evident in patients affected by the congenital variant of RTT, caused by FOXG1 mutations, as well as in classic RTT, thus suggesting a potential contribution for these genes to the behavioral phenotype of RTT.
Interestingly, an altered expression was found for a group of genes (italic in Table 1) expressed in the basal ganglia 29, 30, 31 and involved in the control of movements, some of which are decreased in another developmental disorder, Tourette syndrome.32, 33 Analysis by qRT-PCR confirmed the trend toward upregulation for all of these genes, although the alteration was not statistically significant.
A statistically significant overexpression was identified and confirmed by qRT-PCR for Nnat (represented in bold italics in Table 1). Nnat is highly expressed during fetal and neonatal brain development. Its structure suggests a potential involvement in signal transduction but its function is presently poorly understood. A role in the regulation of ion channels during brain development has been suggested. Accordingly, it has been recently reported that Nnat mRNA is translated in hippocampal dendrites in response to blockade of action potentials and NMDA-receptor-dependent synaptic transmission and can adjust dendritic calcium levels by regulating intracellular calcium storage.34 Authors thus proposed that neuronatin may impact synaptic plasticity by modulating dendritic calcium levels, thereby potentially regulating neuronal excitability, receptor trafficking and calcium-dependent signaling. In adult brain, Nnat is predominantly expressed in parvalbumin-positive GABAergic interneurons. Its levels are increased and accumulated as insoluble aggregates in the cortical area of patients with Lafora disease, a progressive and fatal neurodegenerative disease due to variants in Laforin and Malin genes;35 in this condition, Nnat accumulation is accompanied by a dramatic loss of parvalbumin-positive GABAergic interneurons. Accordingly, the ectopic expression of Nnat in cultured neuronal cells results in increased intracellular Ca(2+), endoplasmic reticulum stress, proteasomal dysfunction and cell death.35 In our experiments, Nnat levels appear significantly higher both in Foxg1+/− whole brain and in silenced hippocampal neurons, strongly suggesting that this gene is directly or indirectly regulated by Foxg1.
Considering the specific role of Foxg1 in the hippocampus during postnatal neurogenesis, we also assessed the expression levels of Nnat and Mef2c in primary hippocampal neurons from E18.5 mouse embryos in which Foxg1 was silenced by shRNA. Consistent with data obtained from whole brain, Nnat expression was significantly upregulated also in hippocampal cultures. As in whole brain, a trend toward deregulation was observed for Mef2c; however, the expression change was in the opposite direction compared with Foxg1+/− whole brain. It is difficult at the moment to explain the reasons for such difference. However, hippocampal cultures may not reach the stage of maturation of P30 brains, and it is possible that the gene is subject to different regulatory mechanisms in different brain regions or in different stages of brain development. Moreover, as in both cases the change was borderline, it is also possible that the observed differences represent normal fluctuations in Mef2c levels, not associated to disease. Further analyses will be thus necessary to confirm the significance of the observed Mef2c alterations.
In conclusion, our data suggest that Foxg1 haploinsufficiency in mouse brain alters gene expression and we suggest new Foxg1 target genes potentially involved in disease pathogenesis. It is interesting to note that none of the known genes belonging to Foxg1 network is present in this list. This is, however, not surprising as the majority of studies have focused on Foxg1 function in early in embryonic mouse brain development and different players are probably involved in its network in postnatal tissue. Further experiments on a larger set of samples will be, however, necessary in order to confirm our results.
References
Chahrour M, Zoghbi HY : The story of Rett syndrome: from clinic to neurobiology. Neuron 2007; 56: 422–437.
Hagberg B : Clinical delineation of Rett syndrome variants. Neuropediatrics 1995; 26: 62.
Rolando S : Rett syndrome: report of eight cases. Brain Dev 1985; 7: 290–296.
Hanefeld F : The clinical pattern of the Rett syndrome. Brain Dev 1985; 7: 320–325.
Zappella M : The Rett girls with preserved speech. Brain Dev 1992; 14: 98–101.
Zappella M : The preserved speech variant of the Rett complex: a report of 8 cases. Eur Child Adolesc Psychiatry 1997; 6 (Suppl 1): 23–25.
Ariani F, Hayek G, Rondinella D et al: FOXG1 is responsible for the congenital variant of Rett syndrome. Am J Hum Genet 2008; 83: 89–93.
Artuso R, Mencarelli MA, Polli R et al: Early-onset seizure variant of Rett syndrome: Definition of the clinical diagnostic criteria. Brain Dev 2010; 32: 17–24.
Roesch A, Mueller AM, Stempfl T, Moehle C, Landthaler M, Vogt T : RBP2-H1/JARID1B is a transcriptional regulator with a tumor suppressive potential in melanoma cells. Int J Cancer 2008; 122: 1047–1057.
Manuel M, Martynoga B, Yu T, West JD, Mason JO, Price DJ : The transcription factor Foxg1 regulates the competence of telencephalic cells to adopt subpallial fates in mice. Development 2010; 137: 487–497.
Tao W, Lai E : Telencephalon-restricted expression of BF-1, a new member of the HNF-3/fork head gene family, in the developing rat brain. Neuron 1992; 8: 957–966.
Murphy DB, Wiese S, Burfeind P et al: Human brain factor 1, a new member of the fork head gene family. Genomics 1994; 21: 551–557.
Xuan S, Baptista CA, Balas G, Tao W, Soares VC, Lai E : Winged helix transcription factor BF-1 is essential for the development of the cerebral hemispheres. Neuron 1995; 14: 1141–1152.
Dou CL, Li S, Lai E : Dual role of brain factor-1 in regulating growth and patterning of the cerebral hemispheres. Cereb Cortex 1999; 9: 543–550.
Hanashima C, Shen L, Li SC, Lai E : Brain factor-1 controls the proliferation and differentiation of neocortical progenitor cells through independent mechanisms. J Neurosci 2002; 22: 6526–6536.
Hanashima C, Li SC, Shen L, Lai E, Fishell G : Foxg1 suppresses early cortical cell fate. Science 2004; 303: 56–59.
Martynoga B, Morrison H, Price DJ, Mason JO : Foxg1 is required for specification of ventral telencephalon and region-specific regulation of dorsal telencephalic precursor proliferation and apoptosis. Dev Biol 2005; 283: 113–127.
Muzio L, Mallamaci A : Foxg1 confines Cajal-Retzius neuronogenesis and hippocampal morphogenesis to the dorsomedial pallium. J Neurosci 2005; 25: 4435–4441.
Hebert JM, Fishell G : The genetics of early telencephalon patterning: some assembly required. Nat Rev Neurosci 2008; 9: 678–685.
Shen L, Nam HS, Song P, Moore H, Anderson SA : FoxG1 haploinsufficiency results in impaired neurogenesis in the postnatal hippocampus and contextual memory deficits. Hippocampus 2006; 16: 875–890.
Siegenthaler JA, Tremper-Wells BA, Miller MW : Foxg1 haploinsufficiency reduces the population of cortical intermediate progenitor cells: effect of increased p21 expression. Cereb Cortex 2008; 18: 1865–1875.
Hébert JM, McConnell SK : Targeting of cre to the Foxg1 (BF-1) locus mediates loxP recombination in the telencephalon and other developing head structures. Dev Biol 2000; 222: 296–306.
Falvella FS, Frullanti E, Galvan A et al: The FGFR4 Gly388Arg polymorphism may affect clinical stage of lung cancer patients by modulating the transcriptional profile of normal lung. Int J Cancer 2009; 124: 2880–2885.
Lambert L, Bienvenu T, Allou L et al: MEF2C mutations are a rare cause of Rett or severe Rett-like encephalopathies. Clin Genet 2012; 82: 499–501.
Zweier M, Gregor A, Zweier C et al: Mutations in MEF2C from the 5q14.3q15 microdeletion syndrome region are a frequent cause of severe mental retardation and diminish MECP2 and CDKL5 expression. Hum Mutat 2010; 31: 722–733.
Lukas M, Neumann ID : Oxytocin and vasopressin in rodent behaviors related to social dysfunctions in autism spectrum disorders. Behav Brain Res 2013; 251: 85–94.
Carini LM, Nephew BC : Effects of early life social stress on endocrinology, maternal behavior, and lactation in rats. Horm Behav 2013; 64: 634–641.
Sala M, Braida D, Donzelli A et al: Mice heterozygous for the oxytocin receptor gene (Oxtr(+/-)) show impaired social behaviour but not increased aggression or cognitive inflexibility: evidence of a selective haploinsufficiency gene effect. J Neuroendocrinol 2013; 25: 107–118.
Parent A, Hazrati LN : Functional anatomy of the basal ganglia. I. The cortico-basal ganglia- thalamo-cortical loop. Brain Res Brain Res Rev 1995; 20: 91–127.
Haber SN : Neurotransmitters in the human and nonhuman primate basal ganglia. Hum Neurobiol 1986; 5: 159–168.
McGinty JF : Co-localization of GABA with other neuroactive substances in the basal ganglia. Prog Brain Res 2007; 160: 273–284.
Kataoka Y, Kalanithi PS, Grantz H et al: Decreased number of parvalbumin and cholinergic interneurons in the striatum of individuals with Tourette syndrome. J Comp Neurol 2010; 518: 277–291.
Lennington JB, Coppola G, Kataoka-Sasaki Y et al: Transcriptome Analysis of the Human Striatum in Tourette Syndrome. Biol Psychiatry 2014; S0006-3223: 00551–00554.
Oyang EL, Davidson BC, Lee W, Poon MM : Functional characterization of the dendritically localized mRNA neuronatin in hippocampal neurons. PLoS One 2011; 6: e24879.
Sharma J, Mukherjee D, Rao SN et al: Neuronatin-mediated aberrant calcium signaling and endoplasmic reticulum stress underlie neuropathology in Lafora disease. J Biol Chem 2013; 288: 9482–9490.
Acknowledgements
The work was partially funded by Telethon grant (GGP09117) and by Italian Health Ministry ‘Ricerca finalizzata 2010’ grant (RF-2010-2317597) to AR.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on European Journal of Human Genetics website
Rights and permissions
About this article
Cite this article
Frullanti, E., Amabile, S., Lolli, M. et al. Altered expression of neuropeptides in FoxG1-null heterozygous mutant mice. Eur J Hum Genet 24, 252–257 (2016). https://doi.org/10.1038/ejhg.2015.79
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2015.79