Abstract
Recent genome-wide association studies have identified common variants at multiple loci influencing lung cancer risk. To decipher the genetic basis of the association signals at 3q28, 5p15.33, 6p21.33, 9p21 and 12p13.33, we performed a meta-analysis of data from five genome-wide association studies in populations of European ancestry totalling 12 316 lung cancer cases and 16 831 controls using imputation to recover untyped genotypes. For four of the regions, it was possible to refine the association signal identifying a smaller region of interest likely to harbour the functional variant. Our analysis did not provide evidence that any of the associations at the loci being a consequence of synthetic associations rather than linkage disequilibrium with a common risk variant at these risk loci.
Similar content being viewed by others
Introduction
Lung cancer causes over one million deaths each year worldwide.1, 2 Although principally caused by tobacco smoking, inherited genetic factors are increasingly being recognised to be important in the aetiology of lung cancer; notably, genome-wide association studies (GWAS) in Europeans have consistently identified polymorphic variation at 15q25.1 (CHRNA5-CHRNA3-CHRNB4), 5p15.33 (TERT-CLPTM1) and 6p21.33 (BAT3-MSH5) as determinants of lung cancer risk.3, 4, 5, 6, 7 Additionally, susceptibility loci for lung cancer at 3q28, 6q22.2, 13q12.12, 10q25.2 and 22q12.2 in Asians have been identified using GWAS.8, 9, 10 Recent studies have validated the 3q28 association in Europeans.11, 12
Non-small-cell lung cancer (NSCLC) is the most frequent histological subtype of lung cancer, comprised primarily of adenocarcinoma (AD) and squamous cell carcinoma (SQ). The various lung cancer histologies have different clinical characteristics reflective of differences in their carcinogenesis and molecular profile.13 Perhaps, not surprisingly, there is variability in the genetic effects on lung cancer risk by histology with subtype-specific associations at 5p15.33 (TERT-CLPTM1) for AD14, 15 and at 9p21 (CDKN2A/CDKN2B)16 and 12p13.33 (RAD52)17 for SQ.
The associations identified by GWAS provide novel insights into the development of lung cancer. However, the tag single-nucleotide polymorphisms (tagSNPs) genotyped are generally not strong candidates for causality, and thus elucidating the functional basis of association signals is challenging. One reason for this is that the correlation matrix between tagSNP(s) and functional variant(s) at any locus can be complex. For example, at least two independent risk loci map to 5p15.33,14 and the 6p22.1–p21.31 locus is part of the human leucocyte antigen (HLA) region, which is highly polymorphic.7 Furthermore, it has recently been proposed that many GWAS signals can be a consequence of ‘synthetic associations’, resulting from the combined effect of one or more rare causal variants rather than simply linkage disequilibrium (LD) with a common risk variant.18
Although it can be hard to deconvolute association signals, functional variant discovery is aided by a deeper examination of genetic variation in the LD blocks in which the tagSNPs reside and this benefits greatly from efforts such as the 1000 Genomes Project, which afford discovery of novel variants.12
To decipher the allelic structure underscoring the associations for lung cancer at nine of the previously reported lung cancer risk-associated regions, 3q28, 5p15.33, 6p21.33, 6q22.2, 9p21, 10q25.2, 12p13.33, 13q12.12 and 22q12.2, we performed a meta-analysis of data from five independent GWAS. To maximise recovery of all variants contributing to lung cancer risk at these loci, we imputed untyped variants using 1000 Genome Project data as the reference panel.
Materials and methods
Subjects and data sets
We used GWAS data from five non-overlapping case–control series of Northern European ancestry, which have been previously reported (Supplementary Table 1): the MD Anderson Cancer Center GWAS of NSCLC comprising 1150 cases and 1134 controls;4 the Institute of Cancer Research (ICR) GWAS comprising 1952 cases and 5200 controls;7 the National Cancer Institute GWAS comprising 5713 cases and 5736 controls;15 the International Agency for Research on Cancer (IARC) GWAS comprising 2533 cases and 3791 controls6 and the Harvard GWAS based on 984 cases and 970 controls,19 which were genotyped using either Illumina HumanHap 317, 317+240S, 370Duo, 550, 610 or 1 M arrays (Illumina Inc., San Diego, CA, USA; Supplementary Table 1).
Ethics
All participants provided informed written consent. All studies were reviewed and approved by institutional ethics review committees at the involved institutions.
Quality control
Standard quality control was performed on all scans excluding individuals with low call rate (<90%) and extremely high or low heterozygosity (ie, P<1.0 × 10−4), as well as all individuals evaluated to be of non-European ancestry (using the HapMap version 2 CEU, JPT/CHB and YRI populations as a reference; Supplementary Table 1). For apparent first-degree relative pairs, we removed the control from a case–control pair; otherwise, we excluded the individual with the lower call rate. Quantile–quantile (Q–Q) plots of genome-wide association test statistics showed that there was minimal inflation rendering substantial cryptic population substructure or differential genotype calling between cases and controls unlikely in each GWAS (genomic control inflation factors, λ=1.00–1.05; Supplementary Figure 1). To bring genotype data obtained from different arrays into a common platform from the five GWAS and to recover untyped genotypes, we imputed SNPs using 1000 Genomes Project data as the reference (Supplementary Table 1). Q–Q plots for all SNPs and those restricted to rare SNPs (minor allele frequency (MAF) <1%) after imputation did not show evidence of substantive overdispersion introduced by imputation (λ=0.99–1.06 and 0.82–1.05).
Statistical analysis
Data were imputed for each scan using 1000 Genome Project data (Phase 1 integrated release 3, March 2012) as the reference, using IMPUTE2 (v.2.1.1),20 MaCH (v.1.0)21 or minimac (v.2012.10.3)22 software (Supplementary Table 1). Genotypes were aligned to the positive strand in both imputation and genotyping. Imputation was conducted separately for each scan in which before imputation each GWAS data set was pruned to a common set of SNPs between cases and controls. As previously advocated, we set thresholds for imputation quality to retain both potential common and rare variants for validation.16, 23 Specifically, poorly imputed SNPs defined by an RSQR <0.30 with MaCH or an information measure Is <0.40 with IMPUTE2 were excluded from the analyses. Tests of association between imputed SNPs and lung cancer were performed using SNPTEST (v.2.5),24 ProbABEL,25 MaCH2dat (v.124)21 or glm function in R. Principal components generated using common SNPs were included in the analysis to limit the effects of cryptic population stratification that might cause inflation of test statistics. Classical HLA alleles were imputed using HLA*IMP2, which is a method for imputing classical HLA alleles from SNP data.26, 27
The association between each SNP and lung cancer risk was assessed by the Cochran–Armitage trend test. The possibility of significant hidden population substructure or differential genotype calling between cases and controls in each GWAS was evaluated using Q–Q plots of test statistics. The inflation factor λ was based on the 90% least significant directly typed SNPs.28 Meta-analysis was undertaken using inverse-variance approaches. Odds ratios (ORs) and associated 95% confidence intervals (CIs) were calculated by unconditional logistic regression using R (v.2.6) and PLINK29 (v.1.06) software. Cochran’s Q-statistic to test for heterogeneity and the I2 statistic to quantify the proportion of the total variation due to heterogeneity were calculated.30 To explore the variability in genetic associations according to tumour histology, we derived ORs for all lung cancer, and AD and SQ cancers. Subset-based meta-analyses of AD and SQ lung cancer, stratified by AD and SQ histology, were conducted using the program ASSET.31 All statistical tests were two-sided.
Bioinformatics
LD metrics were calculated based on 5200 controls from Wellcome Trust Case Control Consortium release 2 (WTCCCII) genotyped using Illumina 1.2 M arrays and plotted using SNAP.32 LD blocks were defined on the basis of HapMap recombination rate (cM/Mb) as defined using the Oxford recombination hotspots and on the basis of distribution of CIs defined by Gabriel et al.33 To explore the epigenetic profile of association signals, we used chromatin state segmentation data generated by the ENCODE project. We used HaploReg34 and RegulomeDB35 to examine whether any of the SNPs or their proxies (ie, r2>0.8 in the 1000 Genomes EUR reference panel) annotate putative transcription factor (TF) binding or enhancer elements. We assessed sequence conservation using GERP; GERP scores (−12 to 6, with 6 being indicative of complete conservation) reflect the proportion of substitutions at that site that are rejected by selection compared with the observed substitutions expected under a neutral evolutionary model based on sequence alignment of 34 mammalian species.36
eQTL, meQTL and mutation analysis
To gain insight into the biological basis of each of the risk loci for lung cancer, we firstly performed an expression quantitative trait loci (eQTL) analysis for directly typed SNPs making use of expression data on over 1100 normal lung tissue.37 To explore the relationship between SNP genotype and gene body methylation made use of previously published methylation quantitative trait loci (meQTL) data from the Tumor Cancer Genome Atlas (TCGA) and the EAGLE study37 using sample size-weighted meta-analysis implemented in METAL.38 To examine the somatic mutation frequency of specific genes, we used data from the analysis of SQ and AD lung cancers generated by TCGA and MutSigCV v.1.439 to determine if the gene harbours more non-synonymous mutations than expected by chance given its size, sequence context and mutation rate. As advocated, we imposed a false discovery rate (FDR) of 0.1 as being statistically significant.39
Results
Subjects and definition of genomic regions
We studied five lung cancer GWAS in populations of European ancestry (Supplementary Table 1). After filtering on the basis of prespecified quality-control measures, the five GWAS provided genotypes on 12 316 lung cancer cases and 16 831 controls (Supplementary Table 1).
To explore the possibility of fine mapping the risk loci for lung cancer, which have been identified in Asians, we recovered the SNPs that had been reported to define respective loci: specifically at rs9387478 (6q22.2, hg19 chr6: g.117786180A>C), rs753955 (13q12.12, hg19 chr13: 24293859T>C), rs7086803 (10q25.2, hg19 chr10: g.114498476A>G) and rs17728461 (22q12.2, hg19 chr22: g.30598552C>G). None of these SNPs displayed a significant association with risk of all lung cancer (ie, P>0.05). For rs9387478, the previously reported risk allele showed an association with both AD and SQ albeit at only P-values of 0.01 and 0.03, respectively. In view of these findings, we did not proceed to formal integration of the regions confining our analysis to loci implicated in Europeans.
We defined the haplotype blocks and recombination hotspots containing the tagSNPs previously reported to be associated with lung cancer risk at 3q28 (TP63, rs4488809, hg19 chr3: g.189356261T>C), 5p15.33 (TERT, rs2736100, hg19 chr5: g.1286516T>G and CLPTM1L, rs402710, hg19 chr5: g.1320722C>T), 6p21.33 (rs3117582, hg19 chr6: g.31620520A>C), 9p21 (CDKN2A/CDKN2B, rs1333040, hg19 chr9: g.22083404C>T) and 12p13.33 (RAD52, rs10849605, hg19 chr12: g.1064438T>C). To include the possibility of long-range synthetic associations, we imputed the regions defined by at least 1 Mb region surrounding the tagSNP associated with lung cancer risk at each of the nine loci. Collectively, the six European and three Asian lung cancer risk loci were captured by 12.5 Mb region of the genome.
For analysis of the 6p21.33 association, we considered the major histocompatibility complex (MHC) to be defined by a 4.5 Mb region bordered by the RFP and MLN genes (rs209130, hg19 chr6: g.28867800A>G and rs1547668, hg19 chr6: g.33775446A>G, respectively) at the telomeric and centromeric ends of 6p21.33. For the HLA imputation, we made use of GWAS SNP data for an extended region in 6p21.33 bounded by rs1165196 (hg19 chr6: g.25813150T>C) and rs2772372 (hg19 chr6: g.33427350T>C).
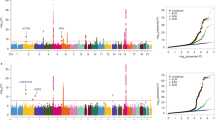
Supplementary Table 1 shows the number of SNPs directly typed and successfully imputed (INFO score ≥0.4) at each of the five loci. Regional plots of association results and recombination rates for all five regions imputed can be found in Figure 1.
Regional plots of association results and recombination rates for 5p15.33 in all lung cancer (a), 3q28 and 5p15.33 in AD (b and c), 6p21.33, 9p21 and 12p13.33 in SQ (d–f). All lung cancer-related panel (a) was based on 12 332 lung cancer cases and 16 831 controls; AD-related panels (b and c) were based on 3930 AD and 15 864 controls; and SQ-related panel (c) was based on 3490 SQ and 16 008 controls. Association results of both genotyped (circles) and imputed (diamonds) SNPs in the GWAS samples and recombination rates for each locus: for each plot, −log10 P-values (y axis) of the SNPs are shown according to their chromosomal positions (x axis). The top genotyped SNP in each combined analysis is a large diamond and is labelled by its rsID. The colour intensity of each symbol reflects the extent of LD with the top genotyped SNP: white (r2=0) through to dark red (r2=1.0). Genetic recombination rates (cM/Mb), estimated using HapMap CEU samples, are shown with a light blue line. Physical positions are based on NCBI build 37 of the human genome. Also shown are the relative positions of genes and transcripts mapping to each region of association. Genes have been redrawn to show the relative positions; therefore, maps are not to physical scale.
Analysis of individual lung cancer risk loci
For four of the five regions, imputation provided refinement of the association signal identifying a region of interest narrower than the original LD block likely to harbour the functional variant. However, for 6p21.33, the LD structure is large and complex. In all regions, in silico functional annotation of the most associated variants, using publically available data from ENCODE, revealed that many reside within potential regulatory regions of DNA.
At 3q28 the association was driven by the relationship with risk for AD (Table 1); the strongest association for this lung cancer histology was provided by rs13314271 (hg19 chr3: g.189357602T>C; P=3.35 × 10−7), which localises to intron 2 of TP63. The strongest eQTL within the region of association was shown by rs4488809 (hg19 chr3: g.189356261T>C) for TP63 (PeQTL=6.77 × 10−14), which is in perfect LD with rs13314271 (D′=1.0, r2=1.0) and resides within a genomic region having epigenetic markers of enhancer function and TF binding (Supplementary Table 3).
The 6p21.33 association is almost exclusively the consequence of an increased risk for SQ lung cancer (Table 1 and Supplementary Table 2). The best association for SQ was provided by rs115549526 (hg19 chr6: g.189356261T>C, P=3.80 × 10−9), which maps to 2.3 kb to 5′ of apolipoprotein M transcript variant 2. In addition to rs115549526, multiple other SNPs within the extended region of LD also show strong associations (ie, within one order of magnitude), which reflect the HLA haplotype structure and the extensive correlations over the whole MHC region. Making use of HLA imputation data, the strongest individual HLA− allelic association for SQ lung cancer was shown by HLA-B*0801 (P=1.09 × 10−9; Supplementary Table 4). Taken together with HLA-A*0101 and DRB*0301, this allele forms the ancestral 8.1 haplotype seen in Europeans. The strongest eQTLs seen in normal lung tissue for the 6p21.33 risk SNPs are shown by rs3131383 (hg19 chr6: g.31704294C>A) and rs497309 (hg19 chr6: g. 31892484T>G) for HLA-A (PeQTL=9.48 × 10−18 and 2.13 × 10−18, respectively) and between rs3117577 (hg19 chr6: g.31727474A>G) and HLA-DQB1 (PeQTL=3.76 × 10−17; Supplementary Table 5). Rs115549526, rs3131383, rs497309 and rs3117577 are all highly correlated SNPs (pairwise LD metrics D′≥0.9, r2≥0.8). The strongest meQTL within the 6p21 risk locus has previously been documented37 to be rs3131379 (hg19 chr6: g.31721033C>T) for MSH5 (PmeQTL=1.14 × 10−17; Supplementary Table 5). Perhaps, not unexpectedly, rs3131379 is strongly correlated with rs115549526 (D′=1.0, r2=0.9). Many of the risk-associated SNPs map to enhancers and have documented TF binding motifs (Supplementary Table 3).
As documented previously,7 two independent associations were shown at 5p15.33, annotating TERT and CLPTM1L genes. The 5p15.33 TERT locus that has been associated with risks of many tumour types with several independent risk loci.40 These risk signals are represented by SNPs in localised regions of LD, within the promoter and within introns 2–4.41, 42 Consistent with previously published observations,15 the strongest 5p15.33–TERT association was provided by rs2736100, which maps to intron 2 of TERT (Figure 1), and is driven by the risk for AD (P=2.51 × 10−18). In contrast, the association defined by rs37004 (hg19 chr5: g.1356684G>A, P=6.91 × 10−16), which maps 12 kb telomeric to CLPTM1L (Figure 1), influenced the risk of both AD and SQ lung cancer histologies. Rs2736100 provided for the strongest meQTL with TERT (PmeQTL=5.28 × 10−19; Supplementary Table 5). For the other 5p15.33 region of association, the intron 13 SNP rs401681 provided the strongest meQTL with CLPTM1L (PmeQTL=7.74 × 10−17; Supplementary Table 5) but was only partially correlated with rs37004 (D′=1.0, r2=0.28).
The signal at 9p21 was primarily driven by risk for SQ, the strongest association being provided by rs1333040 (hg19 chr9: g.22083404C>T), which maps within intron 13 of CDKN2B (P=2.54 × 10−6; Figure 1). Rs1333040 and closely correlated SNPs did not show any significant eQTL or meQTL associations, and did not annotate enhancer or DNAase hypersensitivity elements (Supplementary Table 3).
As expected from previously published data,17 the 12p13.33 association for lung cancer was principally driven by the association with risk of SQ lung cancer; rs3748522 (hg19 chr12: g.1058688A>C) providing the strongest association signal (P=2.35 × 10−8). Rs3748522 maps to intron 1 of the DNA double-strand repair gene RAD52 (Figure 1) within a predicted promoter that features multiple TF binding sites (Supplementary Table 3).
Discussion
Characterizing all the genetic variation within each region of association, as we have performed, is critical in deciphering the allelic architecture responsible for GWAS risk loci and for nominating specific variants for functional analyses. For four of the risk loci, we have been able to refine the association signal identifying a smaller region of interest that is the most likely a location of a functional variant and/or identifying a good candidate.
It is increasingly apparent that multiple independent risk loci for different cancers characterise the 5p15.33 locus. Telomere maintenance is a universal requirement for oncogenic progression. Telomere length (TL) shows substantial interindividual variability and genetically defined TERT-mediated differences in TL represents a promising epidemiological risk factor for cancer. In terms of the TERT risk locus, our findings provide strong evidence that rs2736100 is likely to be responsible for the 5p15.33 association for lung AD. Such an assertion is supported by a recent study demonstrating that rs2736100 genotype is associated with TL variation.43 In contrast, the genetic basis of the CLPTM1L association and risk of all lung cancer is less well defined.
The strongest associations at 3q28 for AD were shown by SNPs mapping to intron 2 of TP63. TP63 is a member of the tumour suppressor TP53 gene family, pivotal to cellular differentiation and responsiveness to cellular stress. High expression levels of TP63 are shown in lung cancers with and without amplification of TP63. Exposure of cells to DNA damage, through carcinogenic agents such as tobacco smoke, leads to induction of TP63 and transactivation of TP53 target genes. As a strong relationship between rs4488809 genotype and TP63 expression was demonstrable, it is entirely plausible that genetically determined differential expression impacts on the ability of impacts on cellular responsiveness to DNA damage. As rs4488809 is one of the top-ranked SNPs and overlaps a predicted enhancer element, it is likely that this SNP represents a tractable basis for the 3q28 association.
As previously stated at 12p13.33, a number of SNPs that are correlated with rs3748522 and map to functional elements could account for this association a priori. In contrast, the 9p21 association is more ill-defined reflecting the more moderate impact of the risk locus in addition to considerations of LD structure. It also worth noting that the Illumina Infinium Humamethylation450 Beadchip (Illumina Inc) used in meQTL analysis has a very low coverage at this locus, which also advocates the level of difficulty in identifying meQTLs in the region.
The 6p21.33 risk locus is large, gene rich and complex. Because it encompasses the major histocompatibility complex, it features many highly correlated variants across a large region making the association signal hard to refine. The meQTL data provides evidence for the role of MSH5 as the basis of the 6p21.33 association. However, a compelling case can be advanced to support HLA variation in defining SQ lung cancer risk. Somatic loss-of-function alterations of HLA-A have been reported previously in genomic studies of lung cancer. Moreover HLA-A is significantly mutated in SQ (FDR=0.07) but not in AD lung cancers. Intriguingly, the observation that HLA-class II variation influences squamous oesophageal cancer risk suggests a wider role for the MHC region in development of solid tumours.
In our study, we found no evidence to support the existence of ‘synthetic associations’ underscoring the currently identified autosomal GWAS signals for lung cancer. At all of the five loci, the variants identified as most associated with lung cancer had an MAF >10%. Although GWAS tagSNPs are unlikely themselves to be functional, they appear much more likely to tag a functional variants of a similar frequency than single or multiple rare causal variants. As some rare variants can be poorly imputed in GWAS, there remains the possibility that some low-frequency variants conferring moderate risks might have been missed. Although inflammatory bowel disease provides support for the existence of ‘synthetic associations’,18 most of the ‘evidence’ for such a model of disease association comes from simulation studies. Indeed, if such a genetic model was present, such associations would be highly tractable by linkage analysis. No putative linkage signals have, however, been identified in these regions.44
Given the caveats in correctly imputing very rare variants, our analysis did not provide evidence that any of the associations at the loci were a consequence of synthetic associations rather than LD with a common risk variant at these risk loci. This is not to say that rare disease-causing variants with large effect do not contribute to the heritable lung cancer risk in general. Evidence for this class of susceptibility is provided by the recent observation that the subpolymorphic variants BRCA2-K3326X and CHEK2-I157T are associated with substantive risk of squamous lung cancer in smokers.12
In summary, we have extensively characterized all genetic variation across five regions that have been reported to be associated with lung cancer in individuals of European decent. In addition to providing insight into the allelic architecture of these association signals our study findings provide a resource informing functional analyses aimed at defining the biological basis of risk loci. Our analysis also reinforces the observation that many of the associations are histology specific. Identifying such histology specific SNPs ultimately should refine our understanding of the origins of morphologic differences, and may contribute to the ongoing search for personalised treatment for subtype-specific lung cancer cases.
References
Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM : Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer 2010; 127: 2893–2917.
Barker K, Martinez A, Wang R et al: PTEN mutations are uncommon in Proteus syndrome. J Med Genet 2001; 38: 480–481.
Hung RJ, McKay JD, Gaborieau V et al: A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature 2008; 452: 633–637.
Amos CI, Wu X, Broderick P et al: Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet 2008; 40: 616–622.
Thorgeirsson TE, Geller F, Sulem P et al: A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature 2008; 452: 638–642.
McKay JD, Hung RJ, Gaborieau V et al: Lung cancer susceptibility locus at 5p15.33. Nat Genet 2008; 40: 1404–1406.
Wang Y, Broderick P, Webb E et al: Common 5p15.33 and 6p21.33 variants influence lung cancer risk. Nat Genet 2008; 40: 1407–1409.
Hu Z, Wu C, Shi Y et al: A genome-wide association study identifies two new lung cancer susceptibility loci at 13q12.12 and 22q12.2 in Han Chinese. Nat Genet 2011; 43: 792–796.
Miki D, Kubo M, Takahashi A et al: Variation in TP63 is associated with lung adenocarcinoma susceptibility in Japanese and Korean populations. Nat Genet 2010; 42: 893–896.
Lan Q, Hsiung CA, Matsuo K et al: Genome-wide association analysis identifies new lung cancer susceptibility loci in never-smoking women in Asia. Nat Genet 2012; 44: 1330–1335.
Wang Y, Broderick P, Matakidou A, Vijayakrishnan J, Eisen T, Houlston RS : Variation in TP63 is associated with lung adenocarcinoma in the UK population. Cancer Epidemiol Biomark Prev 2011; 20: 1453–1462.
Wang Y, McKay JD, Rafnar T et al: Rare variants of large effect in BRCA2 and CHEK2 affect risk of lung cancer. Nat Genet 2014; 46: 736–741.
Travis WD, Brambilla E, Noguchi M et al: International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society: international multidisciplinary classification of lung adenocarcinoma: executive summary. Proc Am Thorac Soc 2011; 8: 381–385.
Broderick P, Wang Y, Vijayakrishnan J et al: Deciphering the impact of common genetic variation on lung cancer risk: a genome-wide association study. Cancer Res 2009; 69: 6633–6641.
Landi MT, Chatterjee N, Yu K et al: A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am J Hum Genet 2009; 85: 679–691.
Timofeeva MN, Hung RJ, Rafnar T et al: Influence of common genetic variation on lung cancer risk: meta-analysis of 14 900 cases and 29 485 controls. Hum Mol Genet 2012; 21: 4980–4995.
Shi J, Chatterjee N, Rotunno M et al: Inherited variation at chromosome 12p13.33, including RAD52, influences the risk of squamous cell lung carcinoma. Cancer Discov 2012; 2: 131–139.
Dickson SP, Wang K, Krantz I, Hakonarson H, Goldstein DB : Rare variants create synthetic genome-wide associations. PLoS Biol 2010; 8: e1000294.
Su L, Zhou W, Asomaning K et al: Genotypes and haplotypes of matrix metalloproteinase 1, 3 and 12 genes and the risk of lung cancer. Carcinogenesis 2006; 27: 1024–1029.
Howie BN, Donnelly P, Marchini J : A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet 2009; 5: e1000529.
Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR : MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol 2010; 34: 816–834.
Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR : Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet 2012; 44: 955–959.
Zeggini E, Scott LJ, Saxena R et al: Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet 2008; 40: 638–645.
Marchini J, Howie B : Genotype imputation for genome-wide association studies. Nat Rev Genet 2010; 11: 499–511.
Aulchenko YS, Struchalin MV, van Duijn CM : ProbABEL package for genome-wide association analysis of imputed data. BMC Bioinform 2010; 11: 134.
Dilthey AT, Moutsianas L, Leslie S, McVean G : HLA*IMP – an integrated framework for imputing classical HLA alleles from SNP genotypes. Bioinformatics 2011; 27: 968–972.
Leslie S, Donnelly P, McVean G : A statistical method for predicting classical HLA alleles from SNP data. Am J Hum Genet 2008; 82: 48–56.
Clayton DG, Walker NM, Smyth DJ et al: Population structure, differential bias and genomic control in a large-scale, case–control association study. Nat Genet 2005; 37: 1243–1246.
Purcell S, Neale B, Todd-Brown K et al: PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81: 559–575.
Higgins JP, Thompson SG, Deeks JJ, Altman DG : Measuring inconsistency in meta-analyses. BMJ 2003; 327: 557–560.
Bhattacharjee S, Rajaraman P, Jacobs KB et al: A subset-based approach improves power and interpretation for the combined analysis of genetic association studies of heterogeneous traits. Am J Hum Genet 2012; 90: 821–835.
Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O'Donnell CJ, de Bakker PI : SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics 2008; 24: 2938–2939.
Gabriel SB, Schaffner SF, Nguyen H et al: The structure of haplotype blocks in the human genome. Science 2002; 296: 2225–2229.
Ward LD, Kellis M : HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic acids Res 2012; 40: D930–D934.
Boyle AP, Hong EL, Hariharan M et al: Annotation of functional variation in personal genomes using RegulomeDB. Genome Res 2012; 22: 1790–1797.
Cooper GM, Stone EA, Asimenos G et al: Distribution and intensity of constraint in mammalian genomic sequence. Genome Res 2005; 15: 901–913.
Shi J, Marconett CN, Duan J et al: Characterizing the genetic basis of methylome diversity in histologically normal human lung tissue. Nat Commun 2014; 5: 3365.
Willer CJ, Li Y, Abecasis GR : METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010; 26: 2190–2191.
Lawrence MS, Stojanov P, Polak P et al: Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 2013; 499: 214–218.
Fletcher O, Houlston RS : Architecture of inherited susceptibility to common cancer. Nat Rev Cancer 2010; 10: 353–361.
Kote-Jarai Z, Saunders EJ, Leongamornlert DA et al: Fine-mapping identifies multiple prostate cancer risk loci at 5p15, one of which associates with TERT expression. Hum Mol Genet 2013; 22: 2520–2528.
Bojesen SE, Pooley KA, Johnatty SE et al: Multiple independent variants at the TERT locus are associated with telomere length and risks of breast and ovarian cancer. Nat Genet 2013; 45: 371–384.
Walsh KM, Codd V, Smirnov IV et al: Variants near TERT and TERC influencing telomere length are associated with high-grade glioma risk. Nat Genet 2014; 46: 731–735.
Bailey-Wilson JE, Amos CI, Pinney SM et al: A major lung cancer susceptibility locus maps to chromosome 6q23–25. Am J Hum Genet 2004; 75: 460–474.
Acknowledgements
This study was supported by grants from the National Institute of Health (NIH) (U19CA148127, 5R01CA055769, 5R01CA127219, 5R01CA133996, and 5R01CA121197, R01 CA111703 and UO1 CA63673, R01 CA092039, CA074386, CA092824, CA090578), Cancer Research UK (C1298/A8780, C1298/A8362), HEAL, Sanofi-Aventis, Institut National du Cancer, the European Community (Integrated Project DNA repair, LSHG-CT-2005-512113), the Norwegian Cancer Association, the Czech Republic (RECAMO, CZ.1.05/2.1.00/03.0101), the Fred Hutchinson Cancer Research Center, an FP7 grant (REGPOT 245536), the Estonian Government (SF0180142s08), by EU RDF in the frame of Centre of Excellence in Genomics and Estoinian Research Infrastructure's Roadmap. We thank all individuals who participated in this study. The ICR study made use of data from the WTCCCII (http://www.wtccc.org.uk). We acknowledge the TCGA for their contribution of lung cancer genomic data to this study (Project Number 3230).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on European Journal of Human Genetics website
Rights and permissions
About this article
Cite this article
Wang, Y., Wei, Y., Gaborieau, V. et al. Deciphering associations for lung cancer risk through imputation and analysis of 12 316 cases and 16 831 controls. Eur J Hum Genet 23, 1723–1728 (2015). https://doi.org/10.1038/ejhg.2015.48
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2015.48
This article is cited by
-
Gene–gene interaction of AhRwith and within the Wntcascade affects susceptibility to lung cancer
European Journal of Medical Research (2022)
-
Large-scale association analysis identifies new lung cancer susceptibility loci and heterogeneity in genetic susceptibility across histological subtypes
Nature Genetics (2017)