Abstract
Causative variants in APP, PSEN1 or PSEN2 account for a majority of cases of autosomal dominant early-onset Alzheimer disease (ADEOAD, onset before 65 years). Variant detection rates in other EOAD patients, that is, with family history of late-onset AD (LOAD) (and no incidence of EOAD) and sporadic cases might be much lower. We analyzed the genomes from 264 patients using whole-exome sequencing (WES) with high depth of coverage: 90 EOAD patients with family history of LOAD and no incidence of EOAD in the family and 174 patients with sporadic AD starting between 51 and 65 years. We found three PSEN1 and one PSEN2 causative, probably or possibly causative variants in four patients (1.5%). Given the absence of PSEN1, PSEN2 and APP causative variants, we investigated whether these 260 patients might be burdened with protein-modifying variants in 20 genes that were previously shown to cause other types of dementia when mutated. For this analysis, we included an additional set of 160 patients who were previously shown to be free of causative variants in PSEN1, PSEN2 and APP: 107 ADEOAD patients and 53 sporadic EOAD patients with an age of onset before 51 years. In these 420 patients, we detected no variant that might modify the function of the 20 dementia-causing genes. We conclude that EOAD patients with family history of LOAD and no incidence of EOAD in the family or patients with sporadic AD starting between 51 and 65 years have a low variant-detection rate in AD genes.
Similar content being viewed by others
Introduction
Autosomal-dominant early-onset Alzheimer disease (ADEOAD) is a rare form of AD affecting patients before the age of 65 years (EOAD) and inherited with an autosomal-dominant pattern. Causative variants (ie, variants reported to cause EOAD with high penetrance when found in a heterozygous state in autosomal-dominant families) in PSEN1, PSEN2, APP (exon 16 or 17) or duplications encompassing the APP locus were found in 77% of families with at least two first-degree relatives from two generations diagnosed with EOAD in our series1 (named thereafter ADEOAD). Some EOAD patients belong to families where one or more affected relatives suffered from AD after 65 years only (late-onset AD, LOAD) and are therefore not diagnosed with ADEOAD. Following our guidelines for AD genetic diagnosis, these patients are not genetically screened. Several other patients present sporadic EOAD. In such cases, genetic screening is less effective. Indeed, following our guidelines for AD genetic diagnosis, we performed genetic screening in patients with sporadic AD with an age of onset before 51 years and found that 16/89 (17.9%) of them carried a causative or probably causative variant in APP, PSEN1 or PSEN2 (data from the French national center for young Alzheimer patients). However, variant-detection rate in patients with sporadic AD and an onset between 51 and 65 years is unknown.
Targeted massive parallel sequencing (next-generation sequencing (NGS)) of a given, clinically significant gene panel has been shown to be a powerful, cost-effective technique to assess the genetic cause of diseases with a high degree of genetic heterogeneity or with numerous differential diagnoses.2, 3, 4 It is also considered to be time-effective compared with sequential Sanger screening of multiple genes. Such a strategy was recently successfully applied to patients with early-onset dementia,5 where a good sensitivity and a good specificity were found. The identification of previously unidentified causal variants in 2/10 samples previously sent for Sanger sequencing of one or more dementia genes illustrated the potential power of this approach. In a research setting, an alternative to targeted sequencing is to perform whole-exome sequencing (WES) and focus first analyses on the same genes as in the gene panel, saving the other genes for later analyses if no causative or probably causative variant is identified. Indeed, WES allows for the massive parallel sequencing of all protein-coding exons, representing 1.2% of the genome, and encoding the approximately 20 000 genes. WES gives access to about 20 000 coding variants per exome, approximately 1000 of which have a frequency of <1% in variant databases or are private variants. One common technical difference between WES and targeted (gene panel) sequencing by NGS is the depth of coverage, which is often designed to be higher in gene panel sequencing than WES, allowing for more accurate variant calling efficiency and therefore fewer false negatives.4 As WES offers insight into nearly all genes and not only the several with diagnostic value or research interest at a given date, using WES could be effective in a research setting, provided there is a good depth of coverage in genes of interest,2, 4 especially in a group of patients with a rather low expected variant-detection rate in genes of diagnostic value.
In an effort to understand the molecular bases of EOAD, we performed WES with high depth of coverage in 424 carefully selected patients with EOAD, including (i) patients with family history of LOAD and no incidence of EOAD in the family (thereafter referred to as ‘LOAD only’) or with sporadic AD starting between 51 and 65 years (no genetic prescreening) and (ii) patients with ADEOAD or sporadic AD with an onset before 51 years and no causative or probably causative variant in the known AD genes. We first searched for APP, PSEN1 and PSEN2 variants and then analyzed for a list of 20 other dementia-causing genes.
Patients and methods
Patients
Patients were recruited by a French memory clinical network including 24 expert centers. Diagnoses were made according to the NINCDS-ADRDA criteria.6 All patients underwent a comprehensive clinical examination, including personal medical and family history assessment, neurological examination, neuropsychological assessment and neuroimaging. When available, CSF AD biomarkers had to be consistent with an AD profile according to the above criteria. The cutoffs used to define a biochemical AD signature were: Aβ42<550 pg/ml, Tau>350 pg/ml, and P-Tau>60 pg/ml. We also calculated the Tau/Aβ42 ratio and a value >0.52 was considered abnormal.7 Two criteria differing by stringency were used to classify CSF samples as supportive of an AD diagnosis: either (i) all three biomarkers were abnormal or (ii) two out of three biomarkers and the Tau/Aβ42 ratio were abnormal. When patients had negative CSF results, the diagnosis of AD was not retained and they were not included. We defined patients with EOAD as patients with AD and an age of onset before 66 years and LOAD patients with an age of onset after 65 years. We defined familial AD as a patient with AD and positive family history of AD, whatever the age of onset, and sporadic AD as patients with no known family history of AD.
All patients gave informed, written consent for genetic analyses. This study was approved by our ethics committee. All blood samples from patients with EOAD referred to our national reference center for young Alzheimer patients were extracted using the Qiagen DNA Blood Kit (Qiagen, Hilden, Germany). APOE genotyping was performed in all EOAD samples by sequencing.
Following our national guidelines for AD genetic diagnosis in EOAD, we sequenced the entire coding region of PSEN1, PSEN2 and exons 16 and 17 of APP by Sanger method and searched for APP duplication by QMPSF as previously described8 in patients with ADEOAD and in patients with sporadic AD with an age of onset before 51 years.
We then selected for WES 424 EOAD patients distributed as following (Figure 1,Supplementary Table S1): (i) patients who did not fulfill our guidelines for AD genetic diagnosis (n=264), including 90 with at least one affected relative with LOAD but none with EOAD (hereafter referred to as ‘LOAD only’) and 174 with sporadic AD and a disease onset between 51 and 65 years and (ii) patients fulfilling our guidelines for AD genetic diagnosis and no causative variant (n=160, including 107 patients with ADEOAD and 53 patients with sporadic AD and an age of onset <51 years).
Summary of the study: patients selected for WES and count of causative, possibly and probably causative variants in AD and other dementia-causing genes. *Unpublished data from the French national center for young Alzheimer patients. **Wallon et al.1 caus., causative.
A total of 9 patients had a neuropathological confirmation of AD diagnosis and 239 had positive CSF biomarkers. The other 176 were carefully selected, in the absence of CSF analysis, on other criteria, using neuropsychological assessments (evidence of a progressive amnestic syndrome of the hippocampal type associated with another cognitive dysfunction) and evidence of neuronal injury using imaging criteria (AD pattern of a cortical atrophy on magnetic resonance imaging and/or decreased 18fluorodeoxyglucose uptake on positron emission tomography).
Whole-exome sequencing
Exomes were captured using the Agilent Sureselect All Exons Human V4+UTR (n=8) or V5 (n=416) Kits (Agilent technologies, Santa Clara, CA, USA). Final libraries were then sequenced on a HiSeq2000 with paired ends, 100-bp reads performed at two sequencing centers. Reads were mapped to the 1000 Genomes GRCh37 build using BWA 0.7.5a.9 Picard Tools 1.101 was used to flag duplicate reads. We applied GATK for indel realignement, base quality score recalibration and SNPs and indels discovery using the Haplotype Caller across all samples simultaneously according to GATK 3.3 Best Practices recommendations.10 The joint variant calling file (VCF) was annotated with refGene gene regions, SNP effects, functional effect prediction tools, as well as Exome Variant Server (EVS) and 1000 Genomes minor allele frequencies (MAFs) using Annovar (http://www.openbioinformatics.org/annovar/).
The annotated VCF was analyzed as following: we first extracted high-quality exonic and splice site variants with a MAF of <1% in the European-American data set of the EVS and/or the European data set of the 1000 Genomes project for PSEN1, PSEN2 and APP genes (variant nomenclatures refer to NM_000021.3 and NM_000447.2, respectively, for PSEN1 and PSEN2 through the main text). We interpreted the variants using the Human Gene Mutation Database (HGMD, www.hgmd.cf.ac.uk), AD&FTD (www.molgen.ua.ac.be/admutations/) and AlzForum (alzforum.org/mutations) databases and by literature search, and finally classified them following the Guerreiro et al11 algorithm. We submitted the variants classified as causative, probably or possibly causative to the Leiden Open Variation Database (LOVD, http://databases.lovd.nl/shared/genes). If no causative, probably or possibly causative variant was identified in these genes, we again extracted variants with MAF<1% among a list of 20 genes known to cause Mendelian forms of other types of dementia, including the frontotemporal dementia spectrum (n=8, MAPT, GRN, VCP, TREM2, SQSTM1, FUS, TARDBP, CHMP2B), dementia with Lewy bodies (n=3, LRRK2, SNCA, PINK1), vascular dementia (n=3, NOTCH3, HTRA1, COL4A1) and other neurodegenerative diseases (n=6, PRNP, DNMT1, ITM2B, SERPINI1, CSF1R, TYROBP), based on a previously published list5, 12 and literature review (Table 1). Variants were then manually annotated using the Human Gene Mutation Database (HGMD, www.hgmd.cf.ac.uk), AD&FTD (www.molgen.ua.ac.be/admutations/) and AlzForum (alzforum.org/mutations) databases, and when reported in at least one database as causative or probably causative, by literature review on a variant to variant basis. New loss-of-function variants (nonsense, frameshift indels and canonical splice site disruptions) in genes where loss of function was documented as the causative mechanism (eg, GRN) were also considered as probably causative. Intronic ‘splice site’ variants were analyzed if located ±5 bp near each coding exon boundary. Deeper intronic analysis was performed for MAPT and GRN, as causative variants were previously reported more deeply.
Variants finally interpreted as causative, probably or possibly causative were confirmed by Sanger sequencing.
Results
PSEN1 and PSEN2 variants
WES was performed in a total of 424 patients with EOAD. Mean depth of coverage of the bases of interest (exons±5 intronic bases) of PSEN1, PSEN2 and APP genes was 159x, 105x and 136x, respectively. An average of 99.97% of bases of interest was covered by at least 10 reads (see also Supplementary Figures S1–S4 for coverage statistics).
No variant was found in exons 16 and 17 of APP. We found three PSEN1 and one PSEN2 causative, possibly or probably causative variants (Table 2, Supplementary Table 2). None of the patients carrying these variants were previously screened by Sanger sequencing, because they did not fulfill our guidelines for AD genetic diagnosis (4/264, 1.5%). Pathogenicity of the PSEN2 c.715A>G, p.(Met239Val) variant was previously reported (in all three databases) (http://databases.lovd.nl/shared/variants/0000061763). The three PSEN1 variants were previously unreported, to our knowledge. Pathogenicity is, however, probable for two and possible for the third, according to the Guerreiro et al11 algorithm. Indeed, the c.691G>C, p.(Ala231Pro) (http://databases.lovd.nl/shared/variants/0000061760) and the c.1169G>A, p.(Ser390Asn) (http://databases.lovd.nl/shared/variants/0000061761) are located at codons where other missense causative variants were reported (respectively, c.692C>T, p.(Ala231Val), c.691G>A, p.(Ala231Thr) and c.1169G>T, p.(Ser390Ile)), the residues are highly conserved (between PSEN1 and PSEN2 and across species) and are therefore predicted to be damaging by most prediction tools (Supplementary Table S2). Conversely, not every prediction tool identifies c.1309A>G, p.(IleI437Val) (http://databases.lovd.nl/shared/variants/0000061762) as damaging, but it is located on a highly conserved residue (between PSEN1 and PSEN2 and across species) and is just contiguous to the two causative variants c.1306C>A, p.(Pro436Gln) and c.1306C>T, p.(Pro436Ser). It is therefore classified as possibly causative.
The four patients with one of the above-mentioned variants had a disease onset between 53 and 57 years (Table 2). Two of them had a sporadic presentation. The father of the patient carrying the PSEN2 causative variant died at age 48 years of another cause with no history of cognitive decline, leading to a censoring effect. No censoring effect was noted in the family of the patient carrying the PSEN1 c.1309A>G, p.(IleI437Val) variant. As the parents’ DNA was not available for testing, we could not check whether the variant occurred de novo or not. The two PSEN1 probably causative variants were found in patients with positive family history of AD, which was not precise enough to fulfill our guidelines for AD genetic diagnosis. In the family with the c.691G>C, p.(Ala231Pro) variant, the pedigree showed that the father died at a young age, leading to a censoring effect. His own father presented AD (unknown age at onset). In the family with the c.1169G>A, p.(Ser390Asn) variant, the paternal aunt was diagnosed with AD at age 68 years, the age of onset was said to be ~3 years before (DNA not available). Surprisingly, the father of the proband (obligate carrier) died at age 68 years with no history of cognitive decline, but the cause of death, cancer and its treatments could have masked signs of cognitive decline related to EOAD, so that autosomal-dominant inheritance was not suggested by family interview.
Eleven other rare variants were identified within APP, PSEN1 and PSEN2 coding sequences (Supplementary Table 2). The significance of the APP rare variants – all located outside exons 16 and 17 – remains to be determined, although their locations reasonably suffice to exclude a causative role in a Mendelian context. The roles of several rare non-synonymous PSEN1 and PSEN2 variants remain, however, uncertain. In particular, we identified an in-frame deletion of Asp 40 (c.116_118del, p.(39_40del)) in PSEN1 in one patient (sporadic, age of onset (AOO) 51 years, APOE 34 genotype), which was already found in a patient with EOAD who presented with frontal dysfunction signs but with no segregation or functional data.13 The same c.116_118del, p.(39_40del) variant has already been reported with a MAF of 0.02% in the EVS. Another rare variant, c.104G>A, p.(Arg35Gln), has also already been detected in controls (MAF of 0.03% in the EVS) and is predicted benign by most software. Moreover, it did not segregate with AD in one French pedigree with another (cosegregating) causative variant (c.360A>C, p.(Glu120Asp), personal data). Additionally, we detected one new missense PSEN1 variant, c.207A>T, p.(Glu69Asp), in a patient with sporadic EOAD starting at the age of 55 years (APOE genotype: 33). This variant has never been reported, is predicted benign by most software and the residue is not conserved in PSEN2. We classified it as likely not causative. Within PSEN2, two other rare missense variants were detected: c.211C>T, p.(Arg71Trp) (in two patients: (1) sporadic, AOO: 60 years, APOE 33 and (2) familial, AOO 65 years, APOE 34, segregation could not be assessed) and c.389C>T, p.(Ser130Leu) (in three patients: (1) sporadic, AOO: 62 years, APOE 33, (2) familial, AOO: 65 years, APOE 33, segregation could not be assessed, and (3) sporadic, AOO: 51 years, APOE 24). Although already found in patients with LOAD, they were also identified in control databases European American EVS and European 1000 Genomes with a MAF of 0.1 and 0.26%, respectively, (c.389C>T) and of 0.37 and 0.01% (c.211C>T), respectively. Moreover, functional assays14 and segregation analyses15 were not in favor of a causative role in a Mendelian context.
Other dementia genes
We next analyzed variants from a list of 20 genes reported to cause other types of dementia when mutated. Mean depth of coverage was 129 × within the bases of interest from this list (see also Supplementary Figures S1–S4 for coverage statistics). Genes were covered in average by at least 10 reads on 98.4% of bases. Except for HTRA1, where only 84% bases of interest were covered by at least 10 reads, all remaining genes were covered on at least an average of 99.2% of bases of interest by 10 reads.
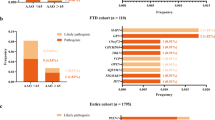
We identified a total of 114 rare non-synonymous variants in 170 patients in the dementia gene list. HGMD classifies variants as ‘DM’ (disease-causing mutation), ‘DM?’ (disease-causing mutation?), DP (disease-associated polymorphism), DFP (disease-associated polymorphism with additional supporting functional evidence) or FP (in vitro/laboratory or in vivo functional polymorphism) according to literature.16 However, falsely classified variants are not rare,17 and this classification is therefore not sufficient to conclude about the pathogenicity of a previously published variant, as illustrated by several variants of unknown significance identified in dementia genes in our data set, classified as ‘DM’ but with evidence against this classification in the literature. Indeed, taking the autosomal-dominant or -recessive inheritance pattern of the selected genes into consideration (Table 1), no patients carried variants that could explain their phenotype. All non-synonymous variants with unknown significance are reported in Supplementary Table S2. We finally could not classify 95 variants (found in 146 patients) in the dementia gene list as causative or not. We cannot exclude the possibility that some of these variants of unknown significance could be reclassified in the future based on putative functional and/or genetic arguments or might confer an increased risk for developing AD.
Discussion
We found that, among the 264 patients not prescreened by Sanger sequencing following our guidelines, only 4 (1.5%) harbored a causative, possibly or probably causative variant within PSEN1 (3) or PSEN2 (1). Censoring effect was observed in the sporadic patient with the PSEN2 variant. The patient carrying the PSEN1 c.1309A>G, p.(IleI437Val) variant had a sporadic presentation of EOAD at age 57 years with no censoring effect, suggesting that, in exceptional cases, sporadic EOAD starting after 50 years may be due to a PSEN1 causative variant. Parents’ DNA was not available, so we could not test the hypothesis of a de novo occurrence.18, 19 Regarding the two patients with positive family history, post hoc analyses of the pedigrees could allow reclassifying them as ADEOAD. Taken together, this suggests that in patients with sporadic EOAD with an age of onset between 51 and 65 years and no censoring effect as well as in patients with EOAD and family history of LOAD only, PSEN1, PSEN2 and APP variant-detection rates might be extremely low, compared with those found with our guidelines for AD genetic diagnosis (ADEOAD and sporadic AD with an age of onset before 51 years). For such patients, WES could be more cost- and time-effective if used as a first-line genetic tool in a research setting. Indeed, only a few patients will be excluded for further research studies regarding the rest of the exome because of a causative PSEN1, PSEN2 or APP variant, while Sanger prescreening of all patients would have taken long time with a high final cost. However, using WES to extract variants from a given gene list in a diagnostic setting should be evaluated in medico-economic studies as it would result in a low rate of results with diagnostic value in this specific group of patients. Conversely, genetic screening by Sanger sequencing (or use of targeted NGS of a gene panel) following our guidelines (ADEOAD and sporadic AD starting before 51) remains a choice strategy, followed by WES in a second line only in variant-negative patients.
In our genetic prescreening, we noted that none of the 11 patients with sporadic AD starting before 51 years and homozygous for the APOE4 genotype carried a causative, possibly or probably causative variant. This was also the case for the five patients selected for WES with sporadic AD starting between 51 and 65 years. APOE4E4 genotype is a strong risk factor for AD and is also considered as a gene with semi-dominant inheritance.20 A larger study focusing on APOE4E4 carriers should be performed to assess the rate of co-occurence with one PSEN1, PSEN2 or APP variant.
The main limitation of our study is that we did not assess copy number variants (CNVs) in the 264 patients with no genetic prescreening (the other patients were prescreened for APP duplications). Of note, APP duplications were previously identified in patients with familial EOAD and/or cerebral amyloid angiopathy with an AOO before 65 years in our series (61 patients from 12 families),1 as well as in the UK series with broader inclusion criteria,21 suggesting that our national guidelines would have allowed identifying all or nearly all APP duplications among the samples referred to our center. APP duplications have sparsely been identified in sporadic cases of early-onset cerebral amyloid angiopathy but were not systematically assessed in EOAD sporadic patients, to our knowledge.22 Further CNV analysis, however, remains necessary.
Importantly, no cause other than AD was found by looking at a list of genes where variants causing Mendelian forms of other types of dementia were reported. Note that we analyzed the TREM2 gene here as a recessively inherited cause of frontotemporal-like dementia. Interestingly, the AD-associated c.140G>A, p.(Arg47His) variant was found in 4 patients (4/424, 0.9%), which is consistent with previous studies.23, 24, 25 In Mendelian diseases, different strategies of WES and whole-genome sequencing allowed the identification of numerous new disease-causing genes and sometimes revealed unexpected results, such as variants affecting function in genes previously known to cause different disorders.26 In addition, NGS allowed enlarging the phenotype associated with several genes, especially in developmental disorders (eg, autism spectrum disorder/intellectual disability/epilepsy)26, 27 and in cancer genetics.26 Regarding neurodegenerative diseases, it is debated whether and how several genes causing well-delineated disorders could be the cause of different neurodegenerative diseases when carrying variants affecting function. For example, C9ORF72 GGGGCC hexanucleotide repeat expansions (which cannot be detected by sequencing), typically causing fronto-temporal dementia – amyotrophic lateral sclerosis spectrum were found in patients with an AD phenotype or Parkinson’s disease, but rarely in controls too, making this finding difficult to interpret.28, 29 It remains possible for these late-onset cases that AD pathology occurred independently of C9ORF72 expansions.30 Similarly, at least one neuropathologically proven AD case was found to harbor a loss-of-function variant of GRN.31 Apparently consistent with the hypothesis of several genes causing different dementing disorders, Guerreiro et al32 reported a CADASIL-causing variant of the NOTCH3 gene in a family clinically diagnosed with AD. However, no evidence of an AD pathophysiological process was available, segregation analysis could not be interpreted, co-occurrence of two distinct diseases could not be excluded as consanguinity was observed and a recessive cause of AD-like dementia remained possible. Jayadev et al33 reported the case of a patient with a nonsense PRNP variant, with a striking amnestic AD-like presentation. Neuropathological examination, however, revealed the presence of PrP immunopositive deposits and no Aβ-positive plaques, eventually confirming that this patient was not affected by AD and that this case was a clinical phenocopy.33 The same nonsense PRNP variant was recently identified by WES in a patient with a similar AD-like clinical presentation, and neuropathological examination was unavailable.34 Use of amyloid biomarkers could have helped reclassify the disease, while clinical history was not suggestive of a classical PrP-associated syndrome. In clinical practice, the diagnosis of AD is usually based on recent McKahnn’s criteria,35 also used in research cohorts36 and international clinical trials.37 Although the sensitivity of these clinical criteria is rather high (80%), their specificity is <70% for probable AD.38, 39 This diagnostic uncertainty has a major impact on molecular diagnostic strategy and may lead to wrongly attribute a disease to a variant affecting function. To help solve this issue, it has been shown that the use of CSF biomarkers can improve the validity of clinical criteria.38, 40, 41, 42 We therefore strongly recommended providing evidence of the AD pathophysiological process prior to any WES.
In conclusion, we identified EOAD patients with no strong predictive arguments for a PSEN1, PSEN2 and APP causative, possibly or probably causative variant as patients with either a positive family history of LOAD only or sporadic EOAD starting after 50 years. The assessment of differential diagnoses by looking at a list of 20 dementia-causing genes in our EOAD patients did not reveal any probably causative variant, suggesting that an enlargement of the phenotype associated with these genes to well-characterized EOAD is unlikely or marginal. However, the role of rare and patient-specific variants among these genes, as well as in AD candidate genes, remains to be elucidated.
References
Wallon D, Rousseau S, Rovelet-Lecrux A et al: The French series of autosomal dominant early onset Alzheimer's disease cases: mutation spectrum and cerebrospinal fluid biomarkers. J Alzheimers Dis 2012; 30: 847–856.
Lohmann K, Klein C : Next generation sequencing and the future of genetic diagnosis. Neurotherapeutics 2014; 11: 699–707.
Della Mina E, Ciccone R, Brustia F et al: Improving molecular diagnosis in epilepsy by a dedicated high-throughput sequencing platform. Eur J Hum Genet 2014; 23: 354–362.
Rehm HL : Disease-targeted sequencing: a cornerstone in the clinic. Nat Rev Genet 2013; 14: 295–300.
Beck J, Pittman A, Adamson G et al: Validation of next-generation sequencing technologies in genetic diagnosis of dementia. Neurobiol Aging 2014; 35: 261–265.
McKhann GM, Knopman DS, Chertkow H et al: The diagnosis of dementia due to Alzheimer's disease: recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement 2011; 7: 263–269.
Duits FH, Prins ND, Lemstra AW et al: Diagnostic impact of CSF biomarkers for Alzheimer's disease in a tertiary memory clinic. Alzheimers Dement 2014; 11: 523–532.
Rovelet-Lecrux A, Hannequin D, Raux G et al: APP locus duplication causes autosomal dominant early-onset Alzheimer disease with cerebral amyloid angiopathy. Nat Genet 2006; 38: 24–26.
Li H, Durbin R : Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010; 26: 589–595.
McKenna A, Hanna M, Banks E et al: The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010; 20: 1297–1303.
Guerreiro RJ, Baquero M, Blesa R et al: Genetic screening of Alzheimer's disease genes in Iberian and African samples yields novel mutations in presenilins and APP. Neurobiol Aging 2010; 31: 725–731.
Rademakers R, Neumann M, Mackenzie IR : Advances in understanding the molecular basis of frontotemporal dementia. Nat Rev Neurol 2012; 8: 423–434.
Nygaard HB, Lippa CF, Mehdi D, Baehring JM : A novel presenilin 1 mutation in early-onset Alzheimer's disease with prominent frontal features. Am J Alzheimers Dis Other Dement 2014; 29: 433–435.
Walker ES, Martinez M, Brunkan AL, Goate A : Presenilin 2 familial Alzheimer's disease mutations result in partial loss of function and dramatic changes in Abeta 42/40 ratios. J Neurochem 2005; 92: 294–301.
Sleegers K, Roks G, Theuns J et al: Familial clustering and genetic risk for dementia in a genetically isolated Dutch population. Brain 2004; 127: 1641–1649.
Stenson PD, Mort M, Ball EV, Shaw K, Phillips A, Cooper DN : The Human Gene Mutation Database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum Genet 2014; 133: 1–9.
Genome of the Netherlands Consortium: Whole-genome sequence variation, population structure and demographic history of the Dutch population. Nat Genet 2014; 46: 818–825.
Dumanchin C, Brice A, Campion D et al: De novo presenilin 1 mutations are rare in clinically sporadic, early onset Alzheimer's disease cases. French Alzheimer's Disease Study Group. J Med Genet 1998; 35: 672–673.
Portet F, Dauvilliers Y, Campion D et al: Very early onset AD with a de novo mutation in the presenilin 1 gene (Met 233 Leu). Neurology 2003; 61: 1136–1137.
Genin E, Hannequin D, Wallon D et al: APOE and Alzheimer disease: a major gene with semi-dominant inheritance. Mol Psychiatry 2011; 16: 903–907.
McNaughton D, Knight W, Guerreiro R et al: Duplication of amyloid precursor protein (APP), but not prion protein (PRNP) gene is a significant cause of early onset dementia in a large UK series. Neurobiol Aging 2012; 33: 426.e13–426.e21.
Llado A, Grau-Rivera O, Sanchez-Valle R et al: Large APP locus duplication in a sporadic case of cerebral haemorrhage. Neurogenetics 2014; 15: 145–149.
Guerreiro R, Wojtas A, Bras J et al: TREM2 variants in Alzheimer's disease. N Engl J Med 2013; 368: 117–127.
Jonsson T, Stefansson H, Steinberg S et al: Variant of TREM2 associated with the risk of Alzheimer's disease. N Engl J Med 2013; 368: 107–116.
Pottier C, Wallon D, Rousseau S et al: TREM2 R47H variant as a risk factor for early-onset Alzheimer's disease. J Alzheimers Dis 2013; 35: 45–49.
Gilissen C, Hoischen A, Brunner HG, Veltman JA : Unlocking Mendelian disease using exome sequencing. Genome Biol 2011; 12: 228.
de Ligt J, Willemsen MH, van Bon BW et al: Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med 2012; 367: 1921–1929.
Beck J, Poulter M, Hensman D et al: Large C9orf72 hexanucleotide repeat expansions are seen in multiple neurodegenerative syndromes and are more frequent than expected in the UK population. Am J Hum Genet 2013; 92: 345–353.
Kohli MA, John-Williams K, Rajbhandary R et al: Repeat expansions in the C9ORF72 gene contribute to Alzheimer's disease in Caucasians. Neurobiol Aging 2013; 34: 1519.e5–1519.e12.
Galimberti D, Arosio B, Fenoglio C et al: Incomplete penetrance of the C9ORF72 hexanucleotide repeat expansions: frequency in a cohort of geriatric non-demented subjects. J Alzheimers Dis 2014; 39: 19–22.
Perry DC, Lehmann M, Yokoyama JS et al: Progranulin mutations as risk factors for Alzheimer disease. JAMA Neurol 2013; 70: 774–778.
Guerreiro RJ, Lohmann E, Kinsella E et al: Exome sequencing reveals an unexpected genetic cause of disease: NOTCH3 mutation in a Turkish family with Alzheimer's disease. Neurobiol Aging 2012; 33: 1008, e1017-1023.
Jayadev S, Nochlin D, Poorkaj P et al: Familial prion disease with Alzheimer disease-like tau pathology and clinical phenotype. Ann Neurol 2011; 69: 712–720.
Guerreiro R, Bras J, Wojtas A, Rademakers R, Hardy J, Graff-Radford N : A nonsense mutation in PRNP associated with clinical Alzheimer's disease. Neurobiol Aging 2014; 35: 2656, e2613-2656.
McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM : Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer's Disease. Neurology 1984; 34: 939–944.
Petersen RC, Aisen PS, Beckett LA et al: Alzheimer's Disease Neuroimaging Initiative (ADNI): clinical characterization. Neurology 2010; 74: 201–209.
Rinne JO, Brooks DJ, Rossor MN et al: 11C-PiB PET assessment of change in fibrillar amyloid-beta load in patients with Alzheimer's disease treated with bapineuzumab: a phase 2, double-blind, placebo-controlled, ascending-dose study. Lancet Neurol 2010; 9: 363–372.
Schoonenboom NS, Reesink FE, Verwey NA et al: Cerebrospinal fluid markers for differential dementia diagnosis in a large memory clinic cohort. Neurology 2012; 78: 47–54.
Lim A, Tsuang D, Kukull W et al: Clinico-neuropathological correlation of Alzheimer's disease in a community-based case series. J Am Geriatr Soc 1999; 47: 564–569.
Andreasson U, Lautner R, Schott JM et al: CSF biomarkers for Alzheimer's pathology and the effect size of APOE varepsilon4. Mol Psychiatry 2014; 19: 148–149.
Dubois B, Feldman HH, Jacova C et al: Advancing research diagnostic criteria for Alzheimer's disease: the IWG-2 criteria. Lancet Neurol 2014; 13: 614–629.
Mouton-Liger F, Wallon D, Troussiere AC et al: Impact of cerebro-spinal fluid biomarkers of Alzheimer's disease in clinical practice: a multicentric study. J Neurol 2014; 261: 144–151.
Acknowledgements
We are grateful to Tracey Avequin for her help in editing the manuscript. This study was funded by grants from the Clinical Research Hospital Program from the French Ministry of Health (GMAJ, PHRC 2008/067) to Didier Hannequin and Dominique Campion, the CNR-MAJ and the JPND PERADES. The funding sources had no specific roles. CNR-MAJ collaborators: Didier Hannequin, Dominique Campion, David Wallon, Olivier Martinaud, Gaël Nicolas (Centre Hospitalo-Universitaire (CHU), Rouen); Olivier Godefroy (CHU Amiens); Frédérique Etcharry-Bouyx, Valérie Chauviré (CHU Angers); Ludivine Chamard, Eric Berger, Eloi Magnin (CHU Besancon); Jean-Francois Dartigues, Sophie Auriacombe (CHU Bordeaux); Vincent de la Sayette, Fausto Viader (CHU Caen); Dominique Castan (CH Castres), Elsa Dionet (CHU Clermont-Ferrand); Francois Sellal (CH Colmar); Olivier Rouaud, Christel Thauvin (CHU Dijon); Olivier Moreaud, Mathilde Sauvée (CHU Grenoble); Adeline Rollin-Sillaire, Stéphanie Bombois, Marie-Anne Mackowiak, Vincent Deramecourt, Florence Pasquier (CHU Lille); Maïté Formaglio, Hélène Mollion, Isabelle Roullet-Solignac, Alain Vighetto, Bernard Croisile (CHU Lyon); Mira Didic, Olivier Félician, Lejla Koric, Mathieu Ceccaldi (CHU Marseille); Audrey Gabelle, Cecilia Marelli, Jacques Touchon, Pierre Labauge (CHU Montpellier); Thérèse Jonveaux (CHU Nancy); Martine Vercelletto, Claire Boutoleau-Bretonnière (CHU Nantes); Giovanni Castelnovo (CHU Nimes); David Renaud, Philippe Robert (CHU de Nice); Claire Paquet, Julien Dumurgier, Jacques Hugon (CHU Lariboisière, Paris); Agnès Michon, Isabelle Le Ber, Bruno Dubois, Charles Duyckaerts (CHU La Salpêtrière, Paris); Foucauld De Boisgueheneuc (CHU Poitiers); Serge Belliard (CHU Rennes); Serge Bakchine (CHU de Reims); Marie-Odile Barrellon, Bernard Laurent (CHU Saint-Etienne); Frédéric Blanc, Christine Tranchant (CHU Strasbourg); Jérémie Pariente, Michèle Puel (CHU Toulouse); and Caroline Hommet, Karl Mondon (CHU Tours).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on European Journal of Human Genetics website
Rights and permissions
About this article
Cite this article
Nicolas, G., Wallon, D., Charbonnier, C. et al. Screening of dementia genes by whole-exome sequencing in early-onset Alzheimer disease: input and lessons. Eur J Hum Genet 24, 710–716 (2016). https://doi.org/10.1038/ejhg.2015.173
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2015.173
This article is cited by
-
Early-onset Alzheimer’s disease due to novel LDLR gene mutation
Acta Neurologica Belgica (2024)
-
One Train May Hide Another: Two Cases of Co-Occurring Primary Familial Brain Calcification and Alzheimer’s Disease
Journal of Molecular Neuroscience (2024)
-
Penetrance estimation of Alzheimer disease in SORL1 loss-of-function variant carriers using a family-based strategy and stratification by APOE genotypes
Genome Medicine (2022)
-
Differential effects of risk factors on the cognitive trajectory of early- and late-onset Alzheimer’s disease
Alzheimer's Research & Therapy (2021)
-
Impaired SorLA maturation and trafficking as a new mechanism for SORL1 missense variants in Alzheimer disease
Acta Neuropathologica Communications (2021)