Abstract
Facioscapulohumeral muscular dystrophy 1 (FSHD1) is caused by a contraction in the number of D4Z4 repeats on chromosome 4, resulting in relaxation of D4Z4 chromatin causing inappropriate expression of DUX4 in skeletal muscle. Clinical severity is inversely related to the number of repeats. In contrast, FSHD2 patients also have inappropriate expression of DUX4 in skeletal muscle, but due to constitutional mutations in SMCHD1 (structural maintenance of chromosomes flexible hinge domain containing 1), which cause global hypomethylation and hence general relaxation of chromatin. Thirty patients originally referred for FSHD testing were screened for SMCHD1 mutations. Twenty-nine had >11 D4Z4 repeats. SMCHD1 c.1040+1G>A, a pathogenic splice-site variant, was identified in a FSHD1 family with a borderline number of D4Z4 repeats (10) and a variable phenotype (in which a LMNA1 sequence variant was previously described), and SMCHD1 c.2606 G>T, a putative missense variant (p.Gly869Val) with strong in vitro indications of pathogenicity, was identified in a family with an unusual muscular dystrophy with some FSHD-like features. The two families described here emphasise the genetic complexity of muscular dystrophies. As SMCHD1 has a wider role in global genomic methylation, the possibility exists that it could be involved in other complex undiagnosed muscle disorders. Thus far, only 15 constitutional mutations have been identified in SMCHD1, and these two sequence variants add to the molecular and phenotypic spectrum associated with FSHD.
Similar content being viewed by others
Introduction
Facioscapulohumeral muscular dystrophy (FSHD) is the third most common muscular dystrophy after the dystrophinopathies and myotonic dystrophy.1, 2, 3 It is inherited in an autosomal dominant manner, affecting about 1 in 20 000 individuals worldwide.4 FSHD is characterised by progressive muscle weakness involving atrophy of the muscles of the face, upper arm and shoulder girdle. General muscle weakness and atrophy may eventually involve the musculature of the pelvic girdle and foot extensors. Early onset of the disease is usually associated with the development of the most severe forms of the disorder.4 Disease onset is unusual before the age of 10 and the disease is usually penetrant by age 30. The majority of patients develop symptoms during the second decade of life. Both retinal vasculopathy and high-tone deafness may be seen as a part of FSHD.
The FSHD1 locus accounts for 95% of clinical disease and maps to 4q35.5 FSHD1 patients carry a large deletion in the polymorphic D4Z4 macrosatellite repeat array at 4q35 and present with 1–11 repeats whereas non-affected individuals possess 12–150 repeats. An almost identical repeat array is present at 10q26; the high sequence identity between these two arrays has proved to be challenging for molecular diagnosis. Each 3.3-kb D4Z4 unit contains a DUX4 (double homeobox 4) gene that, among others, is activated on contraction of the 4q35 repeat array due to the induction of chromatin remodelling of the 4qter region. A number of 4q subtelomeric sequence variants are now recognised, although FSHD1 only occurs in association with ‘permissive’ haplotypes, each of which is associated with a polyadenylation signal located immediately distal of the last D4Z4 unit.6, 7 The resulting poly-A tail appears to stabilise DUX4 mRNAs transcribed from this most distal D4Z4 unit in FSHD muscle cells. Synthesis of both the DUX4 transcripts and the protein in FSHD muscle cells induces significant cell toxicity.8 DUX4 is a transcription factor that may target several genes that results in a deregulation cascade, which, in turn, inhibits myogenesis, sensitises cells to oxidative stress and induces muscle atrophy, thus epitomising many important molecular features of FSHD.
DUX4 is normally expressed in the germline,9, 10 whereas it is epigenetically repressed in healthy somatic cells including differentiated muscle tissue. FSHD1 is caused by the contraction of D4Z4 array that results in DNA hypomethylation and decreased repressive heterochromatin (chromatin relaxation) in the 4q35 region, leading to ectopic DUX4 expression.
Approximately 5% of FSHD patients do not have a contraction in the D4Z4 array, and were historically ascribed to a putative FSHD2 locus. Whole-exome sequencing identified the SMCHD1 gene as the FSHD2 locus.11 The function of the SMCHD1 protein is to aid in the methylation of large chromosomal regions, including the X chromosome and the D4Z4 array. However, not all FSHD2 patients could be explained by contraction of the D4Z4 repeats or mutations in SMCHD1, implying that an FSHD3 locus exists.
Loss of SMCHD1 activity in FSHD2 patients reduces methylation of the D4Z4 array, allowing the genetic transcription machinery to gain access to the DUX4 gene.11 FSHD2 is caused by co-inheritance of two independent events: an FSHD-permissive chromosome 4 haplotype (necessary for polyadenylation of DUX4 mRNA) and a variant in SMCHD1 that causes D4Z4 hypomethylation, neither of which by itself causes disease.11 The SMCHD1 gene product codes for protein that regulates chromatin repression in a wide variety of organisms.10, 12 Given the wider role of SMCHD1 in regulating methylation, it is possible that SMCHD1 could be a modifier in human diseases.
In this study, we screened 30 patients with FSHD or FSHD-like features, of whom 29 did not exhibit a contraction of D4Z4 and 1 a borderline D4Z4 repeat contraction (10 repeats).
Materials and methods
High molecular weight DNA was available from 30 FSHD patients13 who had been referred to our centre for either research or diagnostic testing. All patients had previously undergone DNA diagnostic testing for FSHD1, and all but one had been identified as carrying >11 4q35-located D4Z4 repeats, as determined by p13E11 hybridisation to EcoRI-digested and EcoRI/BlnI-digested genomic DNA.13, 14 The 4qA-defined and 4qB-defined subtelomeric alleles were also identified as previously reported.13, 14 4q35.2-associated haplotypes were assessed as previously reported.15, 16
D4Z4 methylation analysis
Methylation-sensitive restriction digestion
The methylation level in the 4q-associated proximal D4Z4 tandem repeat was determined using the methylation-sensitive enzyme Fse1.11 Following blotting the band intensities were quantified using ImageJ software (v1.43, National Institutes of Health, Bethesda, MD, USA). Some of the samples were also analysed by pyrosequencing of DR1 region of DUX4.17
RNA analysis
RNA was extracted from lymphocytes and was reverse transcribed to complementary DNA (cDNA). Five microliters of 5 ng/μl cDNA was amplified using 40 cycles and sequenced using standard methods.11 Location of primers used is given in Supplementary Table 1.
Bioinformatic analysis
The disease-causing potential of the missense mutation (NM_015295.2:c.2606G>T: p.Gly869Val) was evaluated using Condel (http://bg.upf.edu/condel/home).18 The output of Condel is based on the consensus of in silico predictions from SIFT (http://sift.jcvi.org),19 Polyphen2 (http://genetics.bwh.harvard.edu/pph2)20 and Mutation Assessor (http://mutationassessor.org).21 Condel is a computational method used to assess whether a missense variant results in an amino-acid substitution that is neutral or deleterious to the respective protein. In addition, evolutionary sequence conservation was assessed using a multiple sequence alignment of 13 orthologous proteins. The assessment of the potential impact of p.Gly869Val on splicing was conducted using MutPred Splice (http://mutdb.org/mutpredsplice)22 and by computing the net change to exonic regulatory elements, for example, loss of exonic splicing enhancers (ESE) and/or gain of exonic splicing silencers (ESS), which may serve to weaken exon definition and promote exon skipping. The set of ESE and ESS motifs considered in this analysis are derived from the NI-ESE and NI-ESS set.
Results
Clinical details
The pedigree of family A and B are shown in Figures 1a and 2a, respectively. Individual clinical findings are summarised in Table 1. Detailed clinical findings are described in the Supplementary Material.
Genetic and epigenetic characterisation of family A. (a) Pedigree. Shaded boxes represent affected individuals. + and − denote the presence and absence of variant. (b) Mutation analysis by Sanger sequencing. (c) Reverse transcription-PCR (RT-PCR): the agarose gel images show RT-PCR fragments from the affected individuals. (d) Methylation analysis: DNA digested with EcoRI and Fse1 and hybrised with probe p13E11. Percent methylation for each sample is shown above each lane.
Genetic and epigenetic characterisation of family B. (a) Pedigree. Shaded boxes represent affected individuals. + and − denote the presence and absence of variant. (b) Mutation analysis by Sanger sequencing. (c) Reverse transcription-PCR (RT-PCR): the agarose gel images show RT-PCR fragments from the affected individuals. (d) Methylation analysis: DNA digested with EcoRI and Fse1 and hybrised with probe p13E11. Percent methylation for each sample is shown above each lane. (e) Multiple sequence alignment of 13 orthologous protein.
Molecular genetic and bioinformatic analysis
DNA sequencing of SMCHD1 in 30 FSHD patients exhibiting facial muscle weakness and weakness of either upper limb or lower limb or both revealed a novel obligatory donor splice-site variant c.1040+1G>A in FSHD1 family A (Figure 1b), segregating with disease. All the affected individuals in this family carry 10 units of D4Z4 array. This novel splice-site variant has occurred against the permissive background for FSHD1.11 It was predicted in silico to abolish the 5' splice site (wt=0.98, mut=0), and as RNA analysis did not reveal any exon skipping, it can be concluded that nonsense-mediated decay (NMD) had nullified production of mRNA from the mutant allele (Figure 1c), although in the absence of a normal control this test can only considered to be semi-quantitative. In addition, methylation at D4Z4 loci was observed in all of the affected individuals (17, 16 and 16% ) but not in the unaffected (31%) (Figure 1d).
In family B, owing to the limited amount of DNA available from this family, 4qA analysis was not done but SSLP analysis generated 163 and 166 bp alleles in each of the three sisters, which are unlikely to be on the permissive allele.16 In this family, a novel missense variant, c.2606 G>T, p.Gly869Val was identified in the SMCHD1 gene (Figure 2b). The bioinformatic analysis of p.Gly869Val using Condel predicted this variant to be pathogenic (Condel score=0.896).18
By deriving a prediction based on the consensus of these tools, Condel outperforms each method when applied individually. It has been estimated that Condel achieves an accuracy of ∼88–89% in classifying the impact of a missense variant as neutral or deleterious. A computational method should not be used alone to identify a disease-causing variant, but does give supporting evidence when accompanied by other lines of evidence (for example, segregation and in vitro/in vivo functional studies). In addition, the residue where the variant occurs (p.Gly869) is highly conserved (100%) based on the multiple sequence alignment of 13 orthologous proteins investigated here (Figure 2e). This variant (c.2606G>T) is predicted to disrupt splicing with MutPred Splice.22 If this change was responsible for exon skipping (in a proportion of transcripts), this would cause a frame shift of the reading frame, resulting in a transcript with a premature termination codon (PTC). This mis-spliced transcript containing the PTC may therefore be subject to NMD. The limitation of our RT-PCR assay is that in the absence of a reference gene, it is semi-quantitative (Figure 2c). It is also likely that any pathogenic effect of p.Gly869Val is due to the impact of this amino-acid substitution at the protein level. Due to restricted DNA sample from II-1 and II-2 in family B, DNA samples were analysed by pyrosequencing in another clinically accredited diagnostic laboratory.17 DNA samples from II.2 and II.3 with sequence variant also revealed borderline hypomethylation (37%) based on cutoff being 40% (Debbie Smith, personal communication) whereas II-1 lacked hypomethylation (53%) (Figure 2d).
Discussion
The underlying genetic cause of FSHD has proven singularly difficult to elucidate. It took 21 years following the mapping of the disease to 4q35, to find the underlying genetic cause of FSHD16, 7 and two years later, exome sequencing has revealed that mutations in the SMCHD1 gene are the cause of FSHD2.11 To date, only 15 constitutional mutations causative of FSHD2 have been identified in the SMCHD1 gene.11, 23 Additional two variants from the current study will add to the molecular and phenotypic spectrum associated with FSHD.
Affected members of family A have a phenotype that varies between individuals, but is generally of later onset than classical FSHD (Figure 3). All affected individuals carry 10 D4Z4 repeats on the FSHD-permissive haplotype and also have the pathogenic splice-site variant SMCHD1 c.1040+1G>A.
Clinically, FSHD1 cannot be distinguished from FSHD2.24 Although FSHD1 and FSHD2 have different underlying genetic defects, they both appear to be caused by transcriptional derepression of DUX4 in skeletal muscle.11 The contraction of D4Z4 repeats in FSHD1 is associated with partial demethylation of the shortened allele, and relaxation of the local chromatin structure, whereas, in FSHD2, global genomic demethylation is observed. Hence, given this common mechanism it is reasonable to suppose that combined FSHD1- and FSHD2-associated genetic changes may be instrumental in the development of FSHD. Three scenarios are thus suggested by family A:
-
1
The individuals are affected primarily by their borderline short E/B fragment (10 D4Z4 units), with the SMCHD1 sequence variant acting as a modifier,
-
2
the SMCHD1 sequence variant is primarily causative of their condition and the borderline short E/B fragment is acting as a modifier or
-
3
both the borderline short E/B fragment and SMCHD1 sequence variant are acting equally to cause their disease.23
It is quite possible that other families of this nature and even different individuals within them may be best described by any of the three scenarios. This will only become clear with the study of more individuals and families, and so we would wish to encourage this.
Family A is of additional interest because it has been reported to have a sequence variant in the LMNA gene in association with a remarkably variable phenotype but without perfect segregation of the mutation with the phenotype.25 It is now clear that the LMNA mutation is an innocuous polymorphism. This illustrates the complexity of elucidating the underlying cause of genetic conditions and muscle disorders in particular, which is reflected by our finding of a SMCHD1 sequence variant in the affected individuals.
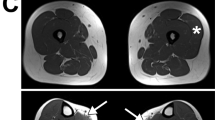
In family B, II.3 (Figure 4) has an undiagnosed muscular dystrophy with an unusual phenotype including some FSHD-like features (facial muscle weakness) and hence why FSHD diagnostic testing was originally requested. However, this individual does not have a short D4Z4 repeat array of the type associated with FSHD1. Results on SSLP analysis identified alleles 163 and 166 in all the three sisters but because of restricted amount of DNA, 4qA analysis was not done. SSLP results suggest that SMCHD1 sequence variant may not have occurred on the permissive background. However, she and her sister (II-2) with sequence change reveal borderline hypomethylation at D4Z4, which would be more compatible with FSHD1 rather than FSHD2. Individual II.4 has not been clinically/molecularly assessed as she is not in the country. The DNA samples from either generation 1 or 3 were not available in this family. FSHD was never considered a likely diagnosis in this family, but testing of FSHD was undertaken to exclude this as a potential confounder. It was intriguing, therefore, to find a novel putative missense change in SMCHD1 (c.2606 G>T, p.Gly869Val) in II.3, which does not appear to segregate with the disease in the family; however, it is possible that II.2 with the variant in this family is non-penetrant for the disease. Bioinformatic analysis strongly suggests the variant to be pathogenic. The amino-acid residue corresponding to human SMCHD1 p.Gly869 is highly conserved in evolution across 13 species, thus implying that it is of functional importance. For confirmation of FSHD, accurate results determining the genetic background on chromosome 4 are necessary in sporadic cases and in patients presenting with an atypical clinical phenotype. Mutations in other genes may also mimic FSHD, including mutations in CAPN3, VCP and FHL1.26, 27, 28 Intriguingly, SMCHD1 c.3651A>G, a missense variant (p.Ile1217Met), has been reported in a family with autistic spectrum disorder,29 which shows that this gene may also have a role to play in non-muscle disorders.
Hence, although family B does not have classical FSHD1 or FSHD2, clinically or molecularly, the possibility exists that SMCHD1 sequence variant may be involved in the pathogenesis of other myopathies and because SMCHD1 has a wider role in global genomic methylation the possibility exists that it could be involved in other complex undiagnosed muscle disorders. This may, of course, be an incidental finding, but certainly suggests further studies defining the role of SMCHD1 in FSHD-associated conditions or other dystrophies are warranted.
References
Upadhyaya M, Cooper DN : Facioscapulohumeral muscular dystrophy clinical medicine and molecular cell biology. Oxon, UK: Garland Science/BIOS Scientific Publishers, 2004.
Van der Maarel SM, Tawil R, Tapscott SJ : Facioscapulohumeral muscular dystrophy and DUX4: breaking the silence. Trends Mol Med 2011; 17: 252–258.
Richards M, Coppée F, Thomas N, Belayew A, Upadhyaya M : Facioscapulohumeral muscular dystrophy (FSHD): an enigma unravelled? Hum Genet 2012; 131: 325–340.
Padberg G : Facioscapulohumeral muscular dystrophy: a clinician’s experience; In Upadhyaya M, Cooper DN (eds). Facioscapulohumeral Muscular Dystrophy Clinical Medicine and Molecular Cell Biology. Oxon, UK: Garland Science/BIOS Scientific Publishers, 2004,, pp 41–53.
Wijmenga C, Hewitt JE, Sandkuijl LA et al: Chromosome 4q DNA rearrangements associated with facioscapulohumeral muscular dystrophy. Nat Genet 1992; 2: 26–30.
Dixit M, Ansseau E, Tassin A et al: DUX4, a candidate gene of facioscapulohumeral muscular dystrophy, encodes a transcriptional activator of PITX1. Proc Natl Acad Sci USA 2007; 104: 18157–18162.
Lemmers RJ, Van der Vliet PJ, Klooster R et al: A unifying genetic model for facioscapulohumeral muscular dystrophy. Science 2010; 329: 1650–1653.
Vanderplanck C, Ansseau E, Charron S et al: The FSHD atrophic myotube phenotype is caused by DUX4 expression. PLoS One 2011; 6: e26820.
Snider L, Geng LN, Lemmers RJ et al: Facioscapulohumeral dystrophy: incomplete suppression of a retrotransposed gene. PLoS Genet 2010; 28 6: e1001181.
Gendrel AV, Tang YA, Suzuki M et al: Smchd1-dependent and -independent pathways determine developmental dynamics of CpG island methylation on the inactive X chromosome. Dev Cell 2012; 23: 265–279.
Lemmers RJ, Tawil R, Petek LM et al: Digenic inheritance of an SMCHD1 mutation and an FSHD-permissive D4Z4 allele causes facioscapulohumeral muscular dystrophy type 2. Nat Genet 2012; 44: 1370–1374.
Blewitt ME, Gendrel AV, Pang Z et al: SmcHD1, containing a structural-maintenance-of-chromosomes hinge domain, has a critical role in X inactivation. Nat Genet 2008; 40: 663–669.
Thomas NST, Wiseman K, Spurlock G, MacDonald M, Üstek D, Upadhyaya M : A large patient study confirming that facioscapulohumeral muscular dystrophy (FSHD) disease expression is almost exclusively associated with an FSHD locus located on a 4qA-defined 4qter subtelomere. J Med Genet 2007; 44: 215–218.
Lemmers RJ, de Kievit P, Sandkuijl L et al: Facioscapulohumeral muscular dystrophy is uniquely associated with one of the two variants of the 4q subtelomere. Nat Genet 2002; 32: 235–236.
Lemmers RJ, Wohlgemuth M, van der Gaag KJ et al: Specific sequence variations within the 4q35 region are associated with facioscapulohumeral muscular dystrophy. Am J Hum Genet 2007; 81: 884–894.
Spurlock G, Jim HP, Upadhyaya M : Confirmation that the specific SSLP microsatellite allele 4qA161 segregates with fascioscapulohumeral muscular dystrophy (FSHD) in a cohort of multiplex and simplex FSHD families. Muscle Nerve 2010; 42: 820–821.
Hartweck LM, Anderson LJ, Lemmers RJ et al: A focal domain of extreme demethylation within D4Z4 in FSHD2. Neurology 2013; 80: 392–399.
González-Pérez A, López-Bigas N : Improving the assessment of the outcome of nonsynonymous SNVs with a consensus deleteriousness score, Condel. Am J Hum Genet 2011; 88: 440–449.
Ng PC, Henikoff S : Predicting deleterious amino acid substitutions. Genome Res 2001; 11: 863–874.
Adzhubei IA, Schmidt S, Peshkin L et al: A method and server for predicting damaging missense mutations. Nat Methods 2008; 7: 248–249.
Reva B, Antipin Y, Sander C : Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res 2011; 39: e118.
Mort M, Sterne-Weiler T, Li B et al: MutPred Splice: machine learning-based prediction of exonic variants that disrupt splicing. Genome Biol 2014; 15: R19.
Sacconi S, Lemmers RJ, Balog J et al: The FSHD2 gene SMCHD1 is a modifier of disease severity in families affected by FSHD1. Am J Hum Genet 2013; 93: 744–751.
De Greef JC, Lemmers RJ, Camano P et al: Clinical features of facioscapulohumeral muscular dystrophy 2. Neurology 2010; 75: 1548–1554.
Rankin J, Auer-Grumbach M, Bagg W et al: Extreme phenotypic diversity and nonpenetrance in families with the LMNA gene mutation R644C. Am J Med Genet A 2008; 146A: 1530–1542.
Angelini C, Nardetto L, Borsato C et al: The clinical course of calpainopathy (LGMD2A) and dysferlinopathy(LGMD2B). Neurol Res 2010; 32: 41–46.
Stojkovic T, Hammouda el H, Richard P et al: Clinical outcome in 19 French and Spanish patients with valosin-containing protein myopathy associated with Paget's disease of bone and frontotemporal dementia. Neuromuscul Disord 2009; 19: 316–323.
Quinzii CM, Vu TH, Min KC et al: X-linked dominant scapuloperoneal myopathy is due to a mutation in the gene encoding four-and-a-half-LIM protein 1. Am J Hum Genet 2008; 82: 208–213.
Neale BM, Kou Y, Liu L et al: Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature 2012; 485: 242–245.
Acknowledgements
We thank patients and their families for their co-operation and Dr R Lemmers for his advice on methylation assays and for providing sequences for the primers for SMCHD1 gene. We also thank Charlotte Philpott for providing technical support. We are most grateful to Debbie Smith and Maggie Williams of the Bristol Genetics Laboratory, North Bristol NHS Trust, for their help with methylation analysis.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Winston, J., Duerden, L., Mort, M. et al. Identification of two novel SMCHD1 sequence variants in families with FSHD-like muscular dystrophy. Eur J Hum Genet 23, 67–71 (2015). https://doi.org/10.1038/ejhg.2014.58
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2014.58
This article is cited by
-
Segregation between SMCHD1 mutation, D4Z4 hypomethylation and Facio-Scapulo-Humeral Dystrophy: a case report
BMC Medical Genetics (2016)
-
Double SMCHD1 variants in FSHD2: the synergistic effect of two SMCHD1 variants on D4Z4 hypomethylation and disease penetrance in FSHD2
European Journal of Human Genetics (2016)
-
Individual epigenetic status of the pathogenic D4Z4 macrosatellite correlates with disease in facioscapulohumeral muscular dystrophy
Clinical Epigenetics (2015)