Abstract
Defects in TRIM32 were reported in limb-girdle muscular dystrophy type 2H (LGMD2H), sarcotubular myopathies (STM) and in Bardet-Biedl syndrome. Few cases have been described to date in LGMD2H/STM, but this gene is not systematically analysed because of the absence of specific signs and difficulties in protein analysis. By using high-throughput variants screening techniques, we identified variants in TRIM32 in two patients presenting nonspecific LGMD. We report the first case of total inactivation by homozygous deletion of the entire TRIM32 gene. Of interest, the deletion removes part of the ASTN2 gene, a large gene in which TRIM32 is nested. Despite the total TRIM32 gene inactivation, the patient does not present a more severe phenotype. However, he developed a mild progressive cognitive impairment that may be related to the loss of function of ASTN2 because association between ASTN2 heterozygous deletions and neurobehavioral disorders was previously reported. Regarding genomic characteristics at breakpoint of the deleted regions of TRIM32, we found a high density of repeated elements, suggesting a possible hotspot. These observations illustrate the importance of high-throughput technologies for identifying molecular defects in LGMD, confirm that total loss of function of TRIM32 is not associated with a specific phenotype and that TRIM32/ASTN2 inactivation could be associated with cognitive impairment.
Similar content being viewed by others
Introduction
Defects in the TRIM32 gene have been reported in two autosomal recessive neuromuscular diseases: limb-girdle muscular dystrophy type 2H (LGMD2H, MIM # 254110)1 and sarcotubular myopathy (STM).2 LGMD2H is a rather mild and progressive myopathy initially described in the Hutterite population, characterized by a wide phenotypic heterogeneity with proximal muscle weakness and wasting, and mildly to moderately increased serum CK levels. The disorder usually occurs within the second or the third decade, the progression is slow and difficulties in ambulation take place at around 40–50 years.3, 4, 5 Pathological examination shows signs of nonspecific muscular dystrophy. STM is a congenital myopathy pathologically characterized by isolated segmental vacuolation of the sarcotubular system.6 It has been shown that both disorders could share common variants.2 Only a few cases of LGMD2H and STM have been reported in the literature and/or in databases (www.lovd.nl/TRIM32) suggesting that these conditions are extremely rare. However, in the absence of reliable commercial antibodies against the protein together with the non-specificity of the clinical presentation, there are no direct or indirect clues pointing to TRIM32 screening (except if patient is of Hutterite colony) and the gene is not routinely screened in diagnostic laboratories.
The TRIM32 gene (NG_011619.1), located on 9q33.1, is composed of two exons with a single coding one (NM_012210.3).1 The gene is ubiquitously expressed and encodes a 72 kDa protein, member of the tripartite motif (TRIM) family possessing an E3 ubiquitin ligase activity.7, 8 TRIM32 contains a RING-finger, a B-box motif, a coiled-coil region and a C terminal domain constituted of six NHL repeats. This last domain is predicted to form a six-bladed beta-propeller structure9 and is known to be involved in protein–protein interactions.1, 10 In particular, TRIM32 interacts with myosin and is able to ubiquitinate actin, suggesting its participation in myofibrillar turnover.10 Of note, the small TRIM32 gene is nested within an intron of the long isoform of the ASTN2 gene (NM_014010.4) which is transcribed from the opposite strand of TRIM32 and exhibits key roles in glial-guided neuronal migration during brain development.11 ASTN2 heterozygous deletions were reported at a high frequency in a number of neurodevelopmental and neurobehavioral conditions such as schizophrenia or autism spectrum disorders.12, 13
The diagnostic process of neuromuscular genetic diseases is tedious and expensive as at least 245 genes have been reported to date (http://www.musclegenetable.fr), including 31 for LGMDs, some of which being very large. High-throughput techniques, that is, CGH (comparative genomic hybridization) array and massively parallel sequencing (MPS), enable searching simultaneously for copy number variations and single nucleotide variations in currently known genes and candidate genes. The EU-funded NMD-Chip project (www.nmd-chip.eu/) was established to design, develop and validate high-throughput DNA approaches to efficiently diagnose patients affected with neuromuscular disorders (NMD) and to identify novel candidate genes implicated in NMD. In this context, 50 genes known to be involved in NMD and hundreds of candidate genes were screened by both CGH arrays and MPS.14, 15 Thanks to these combined approaches, we report here the detection of homozygous and compound heterozygous deletions in TRIM32 in two LGMD patients. Interestingly, the 5’ boundaries of both deletions are located within genomic repeats, signaling possible hotspot for genomic rearrangements and raising the hypothesis that TRIM32 deletions may be more frequent than thought.
Materials and methods
Custom design of the MPS and CGH probes
Custom sets of probes for exonic and exon–intron sequences of 50 genes known to be implicated in LGMD, congenital muscular dystrophies and congenital myopathies15 were manufactured by NimbleGen (Madison, WI, USA)15 for the attention of the NMD-Chip Consortium.
Sequence capture and MPS
After standard extraction, genomic DNA samples were fragmented with the use of Covaris ultrasonicator (Covaris, Woburn, MA, USA), ligated to Illumina (San Diego, CA, USA) multiplexing paired-ends adapters, amplified using a polymerase chain reaction assay with the primers containing sequencing barcodes and hybridized to biotin-labeled custom solution-based capture (SureSelect in solution technology, Agilent, Santa Clara, CA, USA). Hybridization was performed at 65 °C for 72 h, and paired-end sequencing was performed on the Illumina HiSeq 2000 platform to provide mean sequence coverage of more than 100 × with more than 81% of the target bases having at least 30 × coverage.
CGH array analysis
A 12-plex CGH custom array (135 000 probes/sub-array) was manufactured by Roche–Nimblegen (Madison, WI, USA). Experimental steps were performed using standard procedure (version 7.0, http://www.nimblegen.com/). Following fluorescent scanning using the NimbleGen MS200 scanner, the arrays were gridded and sample tracking controls checked using NimbleScan v2.6 software. CGH normalized data were analyzed using the CGHweb online analysis (http://compbio.med.harvard.edu/CGHweb)16 and SignalMap software (Roche–NimbleGen) as previously described.17
Quantitative PCR
Quantitative real-time PCR was performed on genomic DNA from the subjects and a control sample (Boleth, ECAC n°88052031) using primers along the genomic region (Table 1). Copy number assay was performed using the SYBR Chemistry for Real-Time PCR (Fast SYBR Green Master Mix Real-Time PCR Master, Applied Biosystems, Foster City, CA, USA), in the 7900HT Fast Sequence Detection System (Applied Biosystems) using the Chr10 snp rs4750217 as internal reference.
Data submission in a public database
We submitted the data (variants, genetic data and phenotypes) from both patients in the Leiden Open Variant Database (www.LOVD.nl/TRIM32).
Results
Clinical presentation and complementary investigations
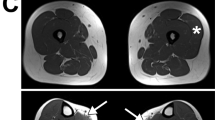
The first patient did not complain of any disability until the age of 30 when he experienced progressive muscle weakness, difficulties in lifting weights, scapular winging and waddling gait. The initial clinical evaluation showed weakness of the glutei, psoas and quadriceps muscles without atrophy, and pointed to a sporadic form of LGMD. When revised 10 years later, the patient was experiencing frequent falls, needed a stick to walk and could neither climb stairs nor tiptoe. CK levels were moderately elevated (400 UI/l, normal range=10–95 UI/l) and electromyography showed a myogenic pattern. Computerized tomography scan showed a rather diffuse damage in the lower limbs with a symmetrical involvement of the posterior thigh muscles (data not shown). The muscle biopsy revealed a non-specific myopathy pattern with a mix of atrophic and hypertrophic fibers, without signs of STM such as vacuolar changes.
The second patient was first seen at the age of 41, but described difficulties to jump or rope climbing since the age of 10 and in climbing stairs since the age of 30. The patient became wheelchair bound at the age of 50. The phenotype was characterized by a pure atrophic presentation with pelvic-femoral predominance. The most affected muscles were the quadriceps, which presented an important atrophy and some contractures. Muscle weakness of the pelvic girdle (gluteus and psoas) and lower limb (hamstring) was also observed associated to areflexia. Even if the patient did not complain of any upper discomfort, symmetrical impairment of biceps, pectoral and supinator longitorum could be noted, the latter one being completely wasted. The upper impairment extended distally later on to the extensorum digitorum (with retractions). No scapular winging, no calf hypertrophy, no facial nor ocular impairment were noted. Of note, over the years, this patient gradually exhibited mild progressive cognitive impairment. He complained of slowness, difficulty to concentrate, omission and fatigability when reading. A mild neuropsychological deficit was confirmed during a neurological examination with a score of 26/30 at the ‘mini-mental state’ test. At the mnemonic level, the patient presented temporal disorientation and weakness of immediate memory. The capacity of logical reasoning and language were preserved. CK levels were normal. The electromyogram was compatible with a myogenic impairment and the muscle computerized tomography scan of the lower limbs showed heterogeneity of the gluteus and of the lower limb muscles with a predominance at the posterior compartment. Although initially normal, the echocardiography at age 50 reported a moderated hypertrophy of the both ventricular walls without left ventricle dysfunction. Simultaneously, the chest X-ray reported a moderate restrictive insufficiency with a forced vital capacity ratio of 80%. A biopsy of the biceps brachialis muscle was performed, showing an aspect compatible with a dystrophic process, without specific signs of STM.
Patients were not from Hutterite origin. No consanguinity or family history of NMD was reported for both patients (Figure 1).
Molecular studies
For patient 1, custom in solution sequence-capture and paired-end MPS was performed on the Illumina platform using the NMD custom libraries generated within the NMD-Chip project.14 A total of 794 genes (3.9 Mb) were sequenced and allowed the identification of an apparent homozygous frameshift mutation in TRIM32 gene, c.1603delC [p.(Leu535Serfs*21)] that was confirmed by Sanger sequencing (Figure 2a and b). Because there was no history of consanguinity, a possible deletion of the other allele of TRIM32 was hypothesized and we thus performed a CGH array analysis. Custom NMD-Chip-CGH-array allowed the identification of a heterozygous 124.4 kb genomic deletion chr9.hg19:g.119447866_119572263del including the whole TRIM32 gene (Figure 2c). For patient 2, a similar CGH approach led to the identification of a homozygous 336 Kb deletion chr9.hg19:g.119447810_119783902del (Figure 2d). The TRIM32 deletion was confirmed at the heterozygous state for patient 1 and homozygous state for patient 2 by quantitative PCR. Unfortunately, DNAs from parents of both patients were not available for parental mutational segregation study.
TRIM32 variants identified (a) IGV viewer visualization of the c.1603delC frameshift mutation appearing as homozygous. (b) Sanger sequencing confirming the homogenous loss of a cytosine. (c) CGH analysis of chromosome 9 allowing the identification of a large deletion encompassing the whole TRIM32 gene in DNA from patient 1. The CGH mean log score value was −0.73 and the deletion covered 242 probes, corresponding to chr9.hg19:g.119447866_119572263del. CGH analysis is visualized with Signalmap software using the unaveraged option (1 × ). (d) CGH analysis of chromosome 9 allowing the identification of a large deletion encompassing the whole TRIM32 gene in DNA from patient 2. The CGH mean log score value was −2.23 and the deletion covered 260 probes, corresponding to chr9.hg19:g.119447810_119783902del.
Examination of the regions around the breakpoints for genomic mapped elements (Human reference sequence (GRCh37) revealed the presence of repetitive elements: two long terminal repeat (LTR) and a long interspersed nuclear elements (LINE), at the location of the proximal breakpoint (Figure 3a). To define more precisely the breakpoints of each deletion, we performed a sequential quantitative PCR analysis by walking along the genomic region (Table 1 and Figure 3a). For both patients, the proximal boundary was located between PCR F15/R15 and F3/R3, defining a region between chr9.hg19:g.119432957 and chr9.hg19:g.119449208 (16 251 bp). Because of the repeated elements, it was not possible to define more precisely the region. The distal boundary of the deletion for patient 1 was defined between PCR F10/R10 and F11/R11, in a region between chr9.hg19:g.119519424 and chr9.hg19:g.119636599 (117 175 bp). For patient 2, the distal boundary of the deletion was defined between the ASTN2 exons 5 and 6: chr9.hg19:g.119770397 and chr9.hg19:g.119802166 (31 769 bp). Of note, both deletions also include, in addition to TRIM32, part of the ASTN2 gene. This gene encodes a 1339 amino acid protein mainly expressed in the brain and characterized by three EGF-like domains and one fibronectin type III domain. Multiple transcript variants have been found for this gene, the largest isoform (NM_014010.4) being composed of 22 exons with TRIM32 being totally included in the intron 15 of this isoform. Interestingly, the defined homozygous deletion in patient 2 would correspond to an out-of-frame deletion because it removes 10 exons from 6 to 15 of the ASTN2 gene corresponding to 1382 nt of the coding sequence (Figure 3).
(a) Analysis of boundaries. Upper panel: In red: primers used for definition of the boundaries. In light green: deletion of patient 1. In dark green: deletion of patient 2. The thinner lines correspond to area of uncertainties. Lower panel: zoom of the repeats present on the upper boundary region. (b) Putative consequences of the homozygous deletion of patient 2 on the ASTN2 protein. From top to bottom: the deletion and the WT ASTN2 protein (c) TRIM32 pathogenic variants reported in the literature and databases (http://www.lovd.nl/TRIM32). Modified from Cossée et al.18
Discussion and Conclusion
So far, only seven different point single nucleotide variations and two heterozygous large deletions were reported in the literature and/or in databases in patients with LGMD2H/STM (www.lovd.nl/TRIM32). All the reported single nucleotide variations are located in the C-terminal part of the protein, in one of the six NHL domains.1, 9, 15, 18, 19 We report here the eighth point mutation in TRIM32. Interestingly, this mutation, p.Leu535Serfs*21, is also located in a NHL domain. As TRIM32 is composed of a single coding exon, it was speculated that the truncating variations are unlikely to destabilize the corresponding mRNA by non-sense mRNA decay. The clustering of single nucleotide variations in the NHL-coding region suggested that these variants may be hypomorphic alleles affecting a function assumed by the NHL domain and do not cause complete loss of function of the TRIM32 protein.18 In these first descriptions, there was no case of TRIM32 deletions. However, some patients were reported as homozygous but a potential deletion on the other allele was not investigated. Since then, two large deletions of the TRIM32 gene have been published.15, 19 Neri et al.15 described, in a 35-year-old patient who experienced progressive muscle weakness, a deletion of the entire TRIM32 gene associated in trans with a nonsense mutation in the last NHL domain. Breakpoints of the deletion were not defined precisely. Borg et al19 reported a family with a complex phenotype of LGMD2H and STM. The patients were compound heterozygotes for a frameshift mutation affecting the fourth NHL domain and a 30 Kb intragenic deletion encompassing parts of TRIM32 intron 1 and the entire exon 2. Interestingly, the patients harbored a pronounced early-onset STM/LGMD2H phenotype compared with previous reported cases. Western blot analysis using homemade antibodies against TRIM32 revealed that no TRIM32 could be detected, suggesting that STM/LGMD2H could be due to a complete loss of function of the protein.19
Here, we report in two patients deletions of the entire TRIM32 gene. The 5’ boundaries of both deletions are located within regions enriched with genomic repeats, suggesting that this region could be prone to rearrangements and that deletions in TRIM32 may be more frequent than initially though. The first patient is compound heterozygous for the deletion and a frameshift mutation. He has a rather mild phenotype of progressive LGMD, as usually described for LGMD2H patients, and the muscle biopsy revealed a non-specific myopathy pattern. The other case is the first description of a homozygous deletion of the whole TRIM32 gene, clinically characterized by an early age of onset compared to classical LGMD2H, but with no evidence of STM on biopsy, normal CK and slow progression with loss of ambulation at the age of 50. Therefore, phenotype of this patient confirms the fact that LGMD2H can be due to a total loss of function of the protein and is not in agreement with a higher severity. Otherwise, this patient gradually developed a mild cognitive impairment. One can hypothesize that the complete loss of function of TRIM32 could affect the cognitive functions as TRIM32 has previously shown to be involved in neuronal differentiation.20, 21, 22, 23 Alternatively, the cognitive impairment may be related to the fact that the deletion also includes part of the ASTN2 gene that codes for astrotactin 2, a brain protein possibly involved in neuronal migration.11 A recent large study revealed that heterozygous copy number variations affecting the 3′-terminal part of ASTN2 or both ASTN2 and TRIM32 are significantly enriched in males with neurodevelopmental and neurobehavioral disorders.13 In this study, the patient with cognitive impairment has a homozygous deletion of ASTN2, whereas the other patient, heterozygous for the ASTN2 deletion, does not present any intellectual disability. This suggests a recessive effect of ASTN2 deletions, with a total loss of function associated with intellectual deficiency. Thereby, one can assume that it could be justified to search for ASTN2/TRIM32 loss of function variants in patients with cognitive impairment.
In conclusion, our results confirm the use of high throughout molecular strategies as a powerful tool to improve the identification of uncovered molecular defect of rare neuromuscular disorders. Identification of new cases of deletions confirm that total loss of function of TRIM32 is not associated with a specific phenotype and illustrates the importance of looking for large deletions in TRIM32, particularly because genomic context suggests the possibility of a recurrent pattern of rearrangements. Finally, our observations suggest that total loss of function of TRIM32/ASTN2 may be associated with cognitive impairment.
References
Frosk P, Weiler T, Nylen E et al: Limb-girdle muscular dystrophy type 2H associated with mutation in TRIM32, a putative E3-ubiquitin-ligase gene. Am J Hum Genet 2002; 70: 663–672.
Schoser BG, Frosk P, Engel AG, Klutzny U, Lochmuller H, Wrogemann K : Commonality of TRIM32 mutation in causing sarcotubular myopathy and LGMD2H. Ann Neurol 2005; 57: 591–595.
Shokeir MH, Kobrinsky NL : Autosomal recessive muscular dystrophy in Manitoba Hutterites. Clin Genet 1976; 9: 197–202.
Shokeir MH, Rozdilsky B : Muscular dystrophy in Saskatchewan Hutterites. Am J Med Genet 1985; 22: 487–493.
Weiler T, Greenberg CR, Nylen E et al: Limb girdle muscular dystrophy in Manitoba Hutterites does not map to any of the known LGMD loci. Am J Med Genet 1997; 72: 363–368.
Jerusalem F, Engel AG, Gomez MR : Sarcotubular myopathy. A newly recognized, benign, congenital, familial muscle disease. Neurology 1973; 23: 897–906.
Reymond A, Meroni G, Fantozzi A et al: The tripartite motif family identifies cell compartments. EMBO J 2001; 20: 2140–2151.
Joazeiro CA, Weissman AM : RING finger proteins: mediators of ubiquitin ligase activity. Cell 2000; 102: 549–552.
Saccone V, Palmieri M, Passamano L et al: Mutations that impair interaction properties of TRIM32 associated with limb-girdle muscular dystrophy 2H. Hum Mutat 2008; 29: 240–247.
Kudryashova E, Kudryashov D, Kramerova I, Spencer MJ : Trim32 is a ubiquitin ligase mutated in limb girdle muscular dystrophy type 2H that binds to skeletal muscle myosin and ubiquitinates actin. J Mol Biol 2005; 354: 413–424.
Wilson PM, Fryer RH, Fang Y, Hatten ME : Astn2, a novel member of the astrotactin gene family, regulates the trafficking of ASTN1 during glial-guided neuronal migration. J Neurosci 2010; 30: 8529–8540.
Wang KS, Liu XF, Aragam N : A genome-wide meta-analysis identifies novel loci associated with schizophrenia and bipolar disorder. Schizophr Res 2010; 124: 192–199.
Lionel AC, Tammimies K, Vaags AK et al: Disruption of the ASTN2/TRIM32 locus at 9q33.1 is a risk factor in males for autism spectrum disorders, ADHD and other neurodevelopmental phenotypes. Hum Mol Genet 2014; 23: 2752–2768.
Bartoli M, Negre P, Wein N et al: Validation of comparative genomic hybridization arrays for the detection of genomic rearrangements of the calpain-3 and dysferlin genes. Clin Genet 2012; 81: 99–101.
Neri M, Selvatici R, Scotton C et al: A patient with limb girdle muscular dystrophy carries a TRIM32 deletion, detected by a novel CGH array, in compound heterozygosis with a nonsense mutation. Neuromuscul Disord 2013; 23: 478–482.
Lai W, Choudhary V, Park PJ : CGHweb: a tool for comparing DNA copy number segmentations from multiple algorithms. Bioinformatics 2008; 24: 1014–1015.
Vasson A, Leroux C, Orhant L et al: Cutom oligonucleotide array-based CGH: a reliable diagnostic tool for detection of exonic copy-number changes in multiple targeted genes. Eur J Human Genet 2013; 21: 977–987
Cossee M, Lagier-Tourenne C, Seguela C et al: Use of SNP array analysis to identify a novel TRIM32 mutation in limb-girdle muscular dystrophy type 2H. Neuromuscul Disord 2009; 19: 255–260.
Borg K, Stucka R, Locke M et al: Intragenic deletion of TRIM32 in compound heterozygotes with sarcotubular myopathy/LGMD2H. Hum Mutat 2009; 30: E831–E844.
Sato T, Okumura F, Iguchi A, Ariga T, Hatakeyama S : TRIM32 promotes retinoic acid receptor alpha-mediated differentiation in human promyelogenous leukemic cell line HL60. Biochem Biophys Res Commun 2012; 417: 594–600.
Hillje AL, Worlitzer MM, Palm T, Schwamborn JC : Neural stem cells maintain their stemness through protein kinase C zeta-mediated inhibition of TRIM32. Stem Cells 2011; 29: 1437–1447.
Gonzalez-Cano L, Hillje AL, Fuertes-Alvarez S et al: Regulatory feedback loop between TP73 and TRIM32. Cell Death Dis 2013; 4: e704.
Hillje AL, Pavlou MA, Beckmann E et al: TRIM32-dependent transcription in adult neural progenitor cells regulates neuronal differentiation. Cell Death Dis 2013; 4: e976.
Acknowledgements
This work was supported by the NMD-Chip project from the European Commission under its seventh framework program (FP7) under the call for ‘High throughput molecular diagnostics in individual patients for genetic diseases with heterogeneous clinical presentation’, N° HEALTH-F5-2008-223026. We thank the Post-Genomic Platform Pitié-Salpêtrière (P3S), Paris, France, for performing MPS.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Nectoux, J., de Cid, R., Baulande, S. et al. Detection of TRIM32 deletions in LGMD patients analyzed by a combined strategy of CGH array and massively parallel sequencing. Eur J Hum Genet 23, 929–934 (2015). https://doi.org/10.1038/ejhg.2014.223
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2014.223
This article is cited by
-
TRIM32 biallelic defects cause limb-girdle muscular dystrophy R8: identification of two novel mutations and investigation of genotype–phenotype correlation
Skeletal Muscle (2023)
-
Altered myogenesis and premature senescence underlie human TRIM32-related myopathy
Acta Neuropathologica Communications (2019)
-
Thin is required for cell death in the Drosophila abdominal muscles by targeting DIAP1
Cell Death & Disease (2018)