Abstract
Recessive nebulin (NEB) mutations are a common cause of nemaline myopathy (NM), typically characterized by generalized weakness of early-onset and nemaline rods on muscle biopsy. Exceptional adult cases with additional cores and an isolated distal weakness have been reported. The large NEB gene with 183 exons has been an obstacle for the genetic work-up. Here we report a childhood-onset case with distal weakness and a core-rod myopathy, associated with recessive NEB mutations identified by next generation sequencing (NGS). This 6-year-old boy presented with a history of gross-motor difficulties following a normal early development. He had distal leg weakness with bilateral foot drop, as well as axial muscle weakness, scoliosis and spinal rigidity; additionally he required nocturnal respiratory support. Muscle magnetic resonance (MR) imaging showed distal involvement in the medial and anterior compartment of the lower leg. A muscle biopsy featured both rods and cores. Initial targeted testing identified a heterozygous Nebulin exon 55 deletion. Further analysis using NGS revealed a frameshifting 4 bp duplication, c.24372_24375dup (P.Val8126fs), on the opposite allele. This case illustrates that NEB mutations can cause childhood onset distal NM, with additional cores on muscle biopsy and proves the diagnostic utility of NGS for myopathies, particularly when large genes are implicated.
Similar content being viewed by others
INTRODUCTION
Nemaline myopathy (NM) is a genetically heterogenous congenital myopathy due to mutations in different genes encoding proteins involved in thin filament assembly and function. Mutations in the nebulin (NEB) gene are the most common cause of autosomal-recessive NM. Mutations are notoriously difficult to identify due to the large size of this giant gene, comprising 183 exons spanning 249 kb of genomic sequence. NEB encodes nebulin, a protein of ∼800 KDa.1, 2 Although the precise function of nebulin is not fully understood, observations in gene knock-out animals suggest that it has a role in the regulation of thin filament length and muscle contraction.3
Typical NEB-related NM is of infantile-onset and static or only slowly progressive, affecting the axial, facial, bulbar and proximal limb muscles. Both milder and more severe forms have been recognized including presentations with profound muscle weakness, arthrogryposis and even neonatal death.4, 5, 6 Late-onset presentations with predominantly distal involvement as well as cores and rods on muscle biopsy have been reported in rare cases.7, 8 Most NEB mutations identified to date are unique, however, a recurrent in-frame deletion of NEB exon 55 has been recognized, associated with a form of NM of variable severity.9, 10
Sequencing of the entire NEB is challenging due to its large size and genotype-phenotype correlations have therefore been difficult to establish. For the genetic diagnosis of congenital myopathies associated with mutations in large genes such as NEB, we therefore developed a next generation sequencing (NGS) approach, which includes copy number analysis for deletions and duplications. Applying this approach, we have uncovered compound heterozygous NEB mutations in a patient with unusual clinic-pathological features, which so far had eluded complete genetic resolution, applying conventional approaches.
MATERIALS AND METHODS
Patient
This is a case report of a patient followed at The Dubowitz Neuromuscular Centre. The patient’s family consented for publication of clinical information and photographs. Informed consent from parents was obtained before the molecular genetic diagnostic work-up.
Molecular analysis
Nebulin Ex55 deletion assay
DNA extracted from peripheral blood was subjected to breakpoint-PCR assay for the NEB exon 55 deletion mutation developed by Anderson et al.9 Identical primer sequences were used in a 10 μl PCR reaction using Qiagen Multiplex Mastermix (Qiagen: www.qiagen.com). Heterozygous and homozygous positive controls were kindly donated by Vilma-LottaLehtokari (The Folkhälsan Institute of Genetics, Helsinki, Finland).
SureSelect probe design
A custom SureSelect library was created using Agilent’s eArray design tool (https://earray.chem.agilent.com/earray/). Briefly, genomic coordinates of 56 genes involved in the congenital myopathies and muscular dystrophies were uploaded to eArray to create a custom library for sequence enrichment before Illumina sequencing (see Supplementary Table 1). Tiling 5 × was used to maximize the chance of detecting insertion and deletion mutations and repetitive regions were omitted from the design using repeat masker.
Illumina sequencing
Sequencing libraries were prepared according to Agilent’s SureSelect Target Enrichment for Illumina Paired-End Multiplexed Sequencing (v1.0) protocol. Following SureSelect capture, samples were subject to index tagging according to the same protocol. Two pools of five uniquely indexed samples were created following quantification by qPCR.11 Each pool was sequenced on one lane of an IlluminaGAIIx.
Sequence alignment
Sequence reads were quality filtered, converted to FASTA format and de-multiplexed using the NextGene software package (v2.0) (Soft Genetics:http://www.softgenetics.com/) and subsequently aligned to a preindexed human genome reference sequence (GRCh37/hg19). Quality filtering and alignment settings and metrics are available upon request.
Muscle biopsy
Frozen sections were stained according to standard procedures.12 Fiber typing was demonstrated by immunolabelling of fast and slow isoforms of myosin heavy chain. Analysis by electron microscopy was used for better characterization of ultrastructural abnormalities.12
Muscle magnetic resonance (MR) imaging
Muscle MR of the lower limb was performed using conventional T1 weighted spin echo according to the protocol previously reported.13 Non-contrast images were obtained from pelvis, thighs and legs.
RESULTS
Case presentation
This 6-year-old boy was referred because of a nasal speech, gross motor difficulties and an abnormal gait.
He was delivered at 37 weeks of gestation by vaginal delivery, following a normal pregnancy as the second of non-identical twins. The neonatal period was uneventful. The parents were non-consanguineous Caucasians with no known Jewish ancestry and no family history of neurological or neuromuscular disorders.
Early motor developmental milestones were within normal limits. By the age of 2 years he started climbing stairs but was unable to run or jump. His speech had a distinct nasal quality and was difficult to understand. In addition, from the age of 4 years he had recurrent chest infections requiring hospitalization.
On clinical examination at 6 years of age (Figures 1 a–c), his weight was on the 0.4th centile while his height was on the 50th centile. There was mild inability to close his eyes while range of eye movements was full. Muscle bulk was generally reduced. He had a positive Gowers’ sign and a waddling, high-stepping gait with bilateral foot drop. There was generalized joint laxity except some tightness in the tendon Achilles and iliotibial bands. Testing of muscle strength on the MRC scale showed pronounced weakness in the distal lower limb and axially, with mild to moderate weakness in more proximal muscle groups (schematic table in Figure 1). He also had a rigid spine and a mild scoliosis. Deep tendon reflexes were difficult to elicit. Serum creatine kinase was 57 IU/l (normal range up to 190 IU/l). Electromyography and nerve conduction studies were interpreted as myopathic. His further course was slowly progressive. At 6.5 years he developed respiratory insufficiency and was started on nocturnal non-invasive positive pressure ventilation. At 8 years he underwent percutaneous endoscopic gastrostomy because of persistent poor weight gain.
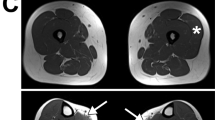
Clinical phenotype and muscle MR imaging at the age of 6 years. (a) General reduction in muscle bulk and unusual feet posture, (b) iliotibial bands tightness and mild rigidity of the spine, (c) mild scoliosis. The table summarizes the testing of muscle strength on the MRC scale. Lower leg muscle MR images (d) the tibialis anterior, peroneal, extensor digitorum and soleus muscles were prominently involved (single arrow) whilst gastrocnemii appeared relatively spared (double arrows). To note also some asymmetry of the anterior compartment involvement with the left lower leg more diffusely involved compared with the right. Picture (e) shows the normal appearance of lower leg muscle MR images in an age-matched control healthy patient.
Histopathology
A muscle biopsy of the quadriceps was performed at the age of 5.5 years showing the presence of both rods and cores, both confirmed with electron microscopy. The relevant histopathology is shown in Figure 2.
Histopathology findings from a quadriceps muscle biopsy performed at 5.5 years. (a) The Gomori trichrome stain showed a wide abnormal variation in fiber size resembling two populations (overall range 5–35 um) and traces of endomysial connective tissue and numerous red staining rods in many fibers of all size. The rods were arranged in peripheral clusters or lines, or diffusely distributed within the fibers. (b) Staining for NADH-TR showed uniform type I fibers and prominent cores with absent enzyme activity in several fibers. The core areas were distinct from the areas with rods that also lacked oxidative enzyme stain. Electron microscopy confirmed the presence of rods (c) and core areas (d) with disrupted myofibrils. Areas with minimal myofibrillar disruption but lacking mitochondria also contributes to the appearance of cores with staining for oxidative enzymes.
Muscle MR imaging
Muscle MR imaging of the lower limbs muscles showed predominant distal involvement. Within the thigh, there were only minor changes in signal intensity without any selective involvement. Within the lower legs, the anterior compartment was more diffusely involved with some asymmetry (details in Figure 1).
Genetic results
Sanger sequencing of the coding exons of the SEPN1, ACTA1, LMNA, RYR1 and KBTBD13 genes did not show any pathogenic mutations. Testing for the known pathogenic NEB exon 55 deletion indicated heterozygosity in the proband and likely association of his condition with the NEB locus.
The proband was screened for mutations in genes involved in congenital muscular dystrophies and myopathies by Illumina sequencing following SureSelect in-solution target enrichment. Sequence variants called by the NextGene software package were filtered, removing variants present in less than 10 percent of reads and greater than 20 base pairs from coding exon boundaries. Further filtering of the remaining 433 variants by absence from dbSNP, specificity to genes targeted in the capture and presence in greater than 20 percent of reads retained 26 variants, including five in NEB (Supplementary Table 2). Of these, a heterozygous four base-pair duplication in exon 172 leading to a frameshift, c.24372_24375dupAAGA (p.Val8126fs), was identified and confirmed by Sanger sequencing. This mutation was described according to HGVS guidelines (http://www.hgvs.org/) relative to reference sequence NM_001271208. 1, where nucleotide number 1 corresponds to the A of the ATG translation initiation codon.
In order to investigate the application of the sequencing protocol to detect large deletions and duplications, log2 ratios of RPKM (Reads Per Kilobase of exon model per Million mapped reads) values were compared for each patient against an average of the other patients for each probe location. Values of ±1 for a heterozygous deletion and ±0.58 for a heterozygous duplication would be expected as deviation from a baseline of 0 using this calculation. We found that the NEB exon 55 deletion in the proband was clearly identifiable using this methodology, with probes in the deleted regions having a ratio of ∼0.8–1.5 against a background of normal probes of ratios typically 0–0.5 (Supplementary Figure 1). We would not however expect to confidently call duplications using this method without further refinement and reduction of the background noise.
We further analyzed the NEB exon 55 deletion by performing a low-stringency alignment (greater than or equal to 40 matching base percentage) of reads rejected from our higher stringency alignments used for variant calling. We were able to identify read pile-ups flanking exon 55 and were thus able to describe the deletion with single base-pair resolution. The deletion in the analyzed patient was identical to that described in Anderson et al.9
By testing the parents we confirmed that the NEB mutations were in trans in the proband; the exon 55 deletion was inherited from the mother and the c.24372_24375dup was inherited from the father.
On the basis of our design all NEB exons were covered sufficiently; the average read depth per exon varied between 169 and 2466, at an average of 577 (Supplementary Table 3).
DISCUSSION
NEB mutations are the most common cause of autosomal-recessive NM but many cases have so far eluded their genetic resolution due to the large size of the NEB gene. Precise genotype-phenotype correlations are currently uncertain.
We have recently introduced NGS into the diagnostic approach to NM and other congenital myopathies. Applying such an approach, we have identified compound heterozygous NEB mutations in a previously unresolved childhood-onset case with unusual clinical and histopathological features, featuring both cores and rods on muscle biopsy and marked distal weakness in the lower limb. A similar clinical phenotype with a childhood-onset, predominant distal myopathy has been recently reported in patients with nemaline rods but no cores on muscle biopsy, due to compound heterozygosity for NEB frameshift and NEB missense or splicing mutations, respectively.14 The combination of cores and rods on muscle biopsy has been described in one isolated NEB-related case, however, this patient was much more severely affected than our case, with hypotonia, profound axial weakness, respiratory impairment from birth and inability to acquire independent ambulation.
Muscle MR images of the lower limb confirmed in our case the predominant distal involvement, however, the anterior compartment was more diffusely involved than has been previously reported in NEB-related NM, where early and isolated involvement of the tibialis anterior muscle is the most typical finding.15, 16
Corresponding to the expanding range of clinical phenotypes associated with NEB mutations, the spectrum of associated radiological features is likely to expand further with increasing efficiency of mutation capture.
Genetic analysis of giant genes such as titin (TTN) and NEB has been very limited to date, as conventional Sanger sequencing of coding exons is extremely time consuming, expensive and only available in few dedicated research centres worldwide. Here we have demonstrated how targeted in-solution hybrid capture systems followed by Illumina NGS has enabled us to genetically resolve an unusual case of a NEB-related myopathy who had eluded genetic resolution applying a more conventional approach. The heterozygous NEB mutation c.24372_24375dup (p.Val8126fs) in exon 172 is predicted to result in a frame shift and a premature stop codon. Donner et al.17 demonstrated that there are four regions of the Nebulin gene which exhibit alternative splicing. Exon 172 is located in the furthest distal of these, exons 166–177, which exhibits an extremely high degree of isoform variability in some tissues. Therefore, isoforms without this mutation would be expected to be present in some muscle tissues, but the proportion of mutant to wild-type transcripts would require further investigation. Additionally, despite this phenomenon, several frameshifting mutations have been described in this region.18
The presence of alternative Nebulin isoforms without the mutated exon 172 could contribute to the clinical variability, and may partially explain the unusual phenotype in this case. Although protein levels are likely to be reduced, we were unable to confirm this due to the large size of the nebulin protein and associated difficulties extracting the nebulin protein from muscle for western blotting. Importantly, we were also able to detect a heterozygous deletion of a single exon 55 by NGS using the log2 ratio of the read counts per probe. This in-frame deletion predicted to result in a reduced protein molecular mass, was initially identified as a founder mutation in the Ashkenazi population but has also recently identified in other populations with less well-defined ancestry.10, 19
These findings demonstrate the strength of targeted NGS as a powerful technique for the detection of point mutations as well as for larger copy number changes.
As currently approximately one third of congenital myopathies remain genetically undiagnosed, our findings demonstrate that NGS is a valuable tool for the molecular diagnosis of congenital myopathies, especially to study large size genes such as NEB.
References
Wang K : Purification of titin and nebulin. Methods Enzymol 1982; 85 ((Pt B):): 264–274.
Donner K, Sandbacka M, Lehtokari VL, Wallgren-Pettersson C, Pelin K : Complete genomic structure of the human nebulin gene and identification of alternatively spliced transcripts. Eur J Hum Genet 2004; 12: 744–751.
Labeit S, Ottenheijm CA, Granzier H : Nebulin, a major player in muscle health and disease. FASEB J 2011; 25: 822–829.
Wallgren-Pettersson C, Donner K, Sewry C et al: Mutations in the nebulin gene can cause severe congenital nemaline myopathy. Neuromuscul Disord 2002; 12: 674–679.
Wallgren-Pettersson C, Pelin K, Nowak KJ et al: Genotype-phenotype correlations in nemaline myopathy caused by mutations in the genes for nebulin and skeletal muscle alpha-actin. Neuromuscul Disord 2004; 14: 461–470.
Lawlor MW, Ottenheijm CA, Lehtokari VL et al: Novel mutations in NEB cause abnormal nebulin expression and markedly impaired muscle force generation in severe nemaline myopathy. Skelet Muscle 2011; 1: 23.
Wallgren-Pettersson C, Lehtokari VL, Kalimo H et al: Distal myopathy caused by homozygous missense mutations in the nebulin gene. Brain 2007; 130 ((Pt 6):): 1465–1476.
Romero NB, Lehtokari VL, Quijano-Roy S et al: Core-rod myopathy caused by mutations in the nebulin gene. Neurology 2009; 73: 1159–1161.
Anderson SL, Ekstein J, Donnelly MC et al: Nemaline myopathy in the Ashkenazi Jewish population is caused by a deletion in the nebulin gene. Hum Genet 2004; 115: 185–190.
Lehtokari VL, Greenleaf RS, DeChene ET et al: The exon 55 deletion in the nebulin gene—one single founder mutation with world-wide occurrence. Neuromuscul Disord 2009; 19: 179–181.
MA Q : A large genome center’s improvements to the Illumina sequencing system. Nat Methods 2008; 5: 1005–1010.
Dubowitz V, Sewry CA : Muscle Biopsy: A Practical Approach 3rd edn Sanders: Elsevier, 2007.
Mercuri E, Pichiecchio A, Counsell S et al: A short protocol for muscle MRI in children with muscular dystrophies. Eur J Paediatr Neurol 2002; 6: 305–307.
Lehtokari VL, Pelin K, Herczegfalvi A et al: Nemaline myopathy caused by mutations in the nebulin gene may present as a distal myopathy. Neuromuscul Disord 2011; 21: 556–562.
Jungbluth H, Davis MR, Müller C et al: Magnetic resonance imaging of muscle in congenital myopathies associated with RYR1 mutations. Neuromuscul Disord 2004; 14: 785–790.
Jungbluth H, Sewry CA, Counsell S et al: Magnetic resonance imaging of muscle in nemaline myopathy. Neuromuscul Disord 2004; 14: 779–784.
Donner K, Sandbacka M, Lehtokari VL et al: Complete genomic structure of the human nebulin gene and identification of alternatively spliced transcripts. Eur J Hum Genet 2004; 12: 744–751.
Pelin K, Donner K, Holmberg M et al: Nebulin mutations in autosomal recessive nemaline myopathy: an update. Neuromuscul Disord 2002; 12: 680–686.
Lehtokari VL, Pelin K, Sandbacka M et al: Identification of 45 novel mutations in the nebulin gene associated with autosomal recessive nemaline myopathy. Human Mutation 2006; 27: 946–956.
Acknowledgements
Thomas Cullup and Heinz Jungbluth were supported by a grant from the Guy’s and St Thomas’ Charitable Foundation (Grant number 070404). The authors acknowledge the use of BRC Core Facilities provided by the financial support from the Department of Health via the National Institute for Health Research (NIHR) comprehensive Biomedical Research Centre award to Guy’s and St Thomas’ NHS Foundation Trust in partnership with King’s College London and King’s College Hospital NHS Foundation Trust. The authors thank the support of the National Commissioning Group to the Dubowitz Neuromuscular Centre for congenital muscular dystrophies and congenital myopathies, and the MRC Neuromuscular Centre Biobank. Francesco Muntoni is funded by Great Ormond Street Hospital Children’s Charity. The authors also wish to thank Vilma-Lotta Lehtokari and Carina Wallgren-Pettersson for kindly donating Nebulin exon 55 deletion control samples.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
All Authors where involved in drafting and reviewing of the manuscript and agreed with the final version for submission. The authors take full responsibility for the data, the analyses and interpretation.
Supplementary Information accompanies this paper on European Journal of Human Genetics website
Rights and permissions
About this article
Cite this article
Scoto, M., Cullup, T., Cirak, S. et al. Nebulin (NEB) mutations in a childhood onset distal myopathy with rods and cores uncovered by next generation sequencing. Eur J Hum Genet 21, 1249–1252 (2013). https://doi.org/10.1038/ejhg.2013.31
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2013.31
Keywords
This article is cited by
-
Self-reported functioning among patients with ultra-rare nemaline myopathy or a related disorder in Finland: a pilot study
Orphanet Journal of Rare Diseases (2023)
-
Ryanodine receptor 1-related disorders: an historical perspective and proposal for a unified nomenclature
Skeletal Muscle (2020)
-
Nebulin nemaline myopathy recapitulated in a compound heterozygous mouse model with both a missense and a nonsense mutation in Neb
Acta Neuropathologica Communications (2020)
-
Bi-allelic mutations in uncoordinated mutant number-45 myosin chaperone B are a cause for congenital myopathy
Acta Neuropathologica Communications (2019)
-
Exome sequencing reveals a nebulin nonsense mutation in a dog model of nemaline myopathy
Mammalian Genome (2016)