Abstract
Our study provides an analysis of the outcome of meiotic segregation of three-way translocations in cleavage-stage embryos and the accuracy and limitations of preimplantation genetic diagnosis (PGD) using the fluorescence in situ hybridization technique. We propose a general model for estimating reproductive risks for carriers of this class of complex chromosome rearrangement. The data presented describe six cycles for four couples where one partner has a three-way translocation. For male heterozygotes, 27.6% of embryos were consistent with 3:3 alternate segregation resulting in a normal or balanced translocation chromosome complement; 41.4% were consistent with 3:3 adjacent segregation of the translocations, comprising 6.9% reflecting adjacent-1 and 34.5% adjacent-2 segregation; 24.1% were consistent with 4:2 nondisjunction; none showed 5:1 or 6:0 segregation; the probable mode could not be ascertained for 6.9% of embryos due to complex mosaicism or nucleus fragmentation. The test accuracy for male heterozygotes was estimated to be 93.1% with 100% sensitivity and 75% specificity. With 72.4% prevalence, the predictive value was estimated to be 91.3% for an abnormal test result and 100% for a normal test result. Two of four couples had a healthy baby following PGD. The proportion of normal/balanced embryo could be significantly less for female heterozygotes, and our model indicates that this could be detrimental to the effectiveness of PGD. A 20% risk of live-born offspring with an unbalanced translocation is generally accepted, largely based on the obstetric history of female heterozygotes; we suggest that a 3% risk may be more appropriate for male carriers.
Similar content being viewed by others
Introduction
Complex chromosomal rearrangements (CCR) are a disparate group1 but the translocations can be generally defined as those involving more than a reciprocal exchange of segments between two chromosomes.2 These can include: (i) two separate simple rearrangements in the same individual (double translocation), (ii) terminal exchanges involving three chromosomes with one breakpoint in each chromosome (three-way translocation), and (iii) more complex rearrangements with multiple breakpoints and often involving different types of rearrangement (terminal exchange, inversion, interstitial insertion, deletion) and more than three chromosomes.3, 4, 5 Constitutional CCRs are very rare and around 255 cases have been described.6
A patient with an apparently balanced de novo CCR and learning difficulties and/or multiple congenital abnormalities may have chromosome imbalance (cryptic or sub-microscopic) associated with the rearranged chromosomes or elsewhere in the genome.7, 8 A person with a normal phenotype who carries a CCR is unlikely to have clinically significant chromosome imbalance; however, the complexity of the CCR may be more extensive than can be seen using karyotyping alone and full characterization is important to assess the risk of progeny with chromosome imbalance; such characterization is essential for accurate prenatal diagnosis.9
Male carriers of CCRs generally present with oligospermia. Testicular biopsy and analysis of spread spermatocytes using electron microscopy has shown that three-way translocations typically form a hexavalent synaptonemal complex at pachytene with a reduced number of chiasmata in the interstitial segments between the centromeres and the breakpoints.10, 11 Although analysis of chromosome complements in mature sperm or cleavage-stage embryos only gives information on the end product of meiosis and not direct observation of the underlying mechanisms, such studies provide useful information on the probable segregation products of the hexavalent complexes at male or female meiosis. This information can be used for risk assessment and counselling as to the value and accuracy of preimplantation genetic diagnosis (PGD).
This paper describes our studies on segregation outcomes for four carriers of three-way translocations and presents a model for comprehensive analysis of these translocations and estimation of the live birth risk of chromosome imbalance for natural pregnancy and the risk reduction following PGD.
Materials and methods
Patients
Our retrospective case series included four couples referred to the Guy’s and St Thomas’ Centre for PGD, three male heterozygotes, and one female with a three-way translocation. Two of the male carriers had oligoasthenoteratozoospermia and one had normozoospermia. None of the couples had a healthy live-born pregnancy before PGD (Table 1).
Case A, 46,XY,t(6;11;16)(q16.2;p14.2;q13): the segment on chromosome 6 distal to 6q16.2 has been translocated onto chromosome 11 at band 11p14.2, the segment on chromosome 11 distal to 11p14.2 onto chromosome 16 at 16q13, and the segment of chromosome 16 distal to 16q13 onto chromosome 6 at 6q16.2.
Case B, 46,XY,t(1;4;14)(p32.3;q23;q13): the segment on chromosome 1 distal to 1p32.3 has been translocated onto chromosome 4 at band 4q23, the segment on chromosome 4 distal to 4q23 onto chromosome 14 at 14q13, and the segment of chromosome 14 distal to 14q13 onto chromosome 1 at 1p32.3.
Case C, 46,XY,t(1;9;18)(p13.3;p22;q23): the segment on chromosome 1 distal to 1p13.3 has been translocated onto chromosome 9 at band 9p22, the segment on chromosome 9 distal to 9p22 onto chromosome 18 at 18q23, and the segment of chromosome 18 distal to 18q23 onto chromosome 1 at 1p13.3.
Case D, 46,XX,t(1;3;4)(q42.1;q26.2;p15.2): the segment on chromosome 1 distal to 1q42.1 has been translocated onto chromosome 3 at band 3q26.2, the segment on chromosome 3 distal to 3q26.2 onto chromosome 4 at 4p15.2, and the segment of chromosome 4 distal to 4p15.2 onto chromosome 1 at 1q42.1.
This report is part of a long-term study of PGD, which was approved by the Guy’s and St Thomas’ Hospital Ethics Committee, Lambeth, Southwark and Lewisham Health Commission (Ref: EC93/046). The PGD cycles included in this study have been reported to the European Society of Human Reproduction and Embryology PGD Consortium annual data collections.
Assisted conception and genetic testing
Procedures were performed as described previously.12, 13 In brief, a standard long stimulation protocol for controlled ovarian stimulation was followed by intracytoplasmic sperm injection for the male heterozygotes due to poor semen characteristics and IVF for the female case, then biopsy of one cell from cleavage-stage embryos 3 days after fertilization and embryo transfer on day 5. Written consent was obtained from the couples for testing and further study of their embryos in accordance with Human Fertilization and Embryology Authority research licence R0075.
Fluorescence in situ hybridization (FISH) probes were directly or indirectly labelled and sourced from different manufacturers (Supplementary Appendix I). A total of five probes were used for each translocation in order to detect all the theoretical unbalanced translocation products (Supplementary Appendix II), one probe for each of the three translocated segments and one probe each for two of the three centric segments. The choice of probes was informed by the fluorophores available, the predicted most frequent products leading to chromosome imbalance, and any products likely to result in live-born offspring with chromosome imbalance.14 Case A had two sequential hybridization reactions of the same nucleus, and Cases B, C, and D a single hybridization. The risk of clinically significant misdiagnosis (a viable unbalanced translocation product with a normal/balanced biopsy result) was minimized by ensuring there were two diagnostic probes for potentially viable imbalance. The probe mixes were tested on cultured lymphocyte metaphase spreads and interphase nuclei from both reproductive partners using standard cytogenetics techniques, as described previously.13 Individual probe efficiencies ranged from 86–99% (Supplementary Appendix I) and the overall suitability of each probe mix was assessed using the predictive value calculated for a normal and abnormal test result (the probability that the result is correct) and an assumed 88% prevalence of chromosome imbalance, with a pass threshold of 95% negative and 85% positive predictive value. Blastomere nuclei from biopsied embryos were hybridized overnight and analysed as described previously.13
Chromosome segment copy number of blastomere nuclei cannot be used to deduce with complete certainty the mode of meiotic segregation of the CCR at pachytene; this is because of meiotic recombination and meiosis II or early cleavage-stage nondisjunction, in addition to possible imbalance contributed by the non-carrier partner. However, accepting the limitations, the probable mode of meiotic segregation based on segment copy number was elucidated as described previously.15, 16 Normal/balanced biopsy results without COD were assigned to be consistent with meiotic 3:3 alternate segregation where the nucleus showed two signals for each chromosome region tested. For the diagnostic accuracy study, spread embryos were confirmed to be normal (balanced) if at least 50% of nuclei were consistent with normal copy number.16, 17 Abnormal tests results were deviations from a normal test result; spread embryos were confirmed to be abnormal if >50% of nuclei were abnormal, and assigned to be consistent with 3:3 adjacent, 4:2, 5:1, or 6:0 disjunction at meiosis if at least two nuclei obtained showed the appropriate signal patterns. The probable segregation mode was deemed to be unknown if these criteria were not met.
Statistical analysis
For the diagnostic accuracy part of the study, biopsy (index) results with an unknown outcome (reference standard) were allocated in proportion to normal and abnormal biopsy results with a known outcome.16, 17 Diagnostic accuracy measures calculated were: false positive and false negative (incorrect abnormal and normal biopsy results using the test perspective and calculated as the proportion of the total outcomes), overall accuracy (the proportion of all biopsy results that were correct), sensitivity (the proportion of abnormal embryos that had an abnormal biopsy result) and specificity (the proportion of normal embryos that had a normal biopsy result), the positive predictive value (the proportion of abnormal biopsy results that were correct) and the negative predictive value (the proportion of normal biopsy results that were correct).
Fisher’s exact test was used to calculate significance probabilities. For study measures, 95% confidence intervals (CIs) were calculated to indicate precision.
Three-way translocation model
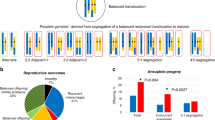
Supplementary Appendix III is a flexible version of the model and includes further technical detail and worked examples using cases A–D. In brief, hexavalent synapsis at meiosis I with 64 segregation products is assumed (see Figure 1). The proportion of each mode for male heterozygotes is based on the spermatozoa study of Pellester et al.18 The proportion of each segregation mode for a female heterozygote was derived using a selection coefficient (s) to reflect checkpoints associated with male meiosis (Table 2). The calculation assumes the same proportion (p) for each product within each mode. The proportion of each mode (∑p) is therefore the number of products for each mode × p. The selection coefficient (s=0.12568) was estimated from the relative proportion of 3:3 alternate (assumed to have 100% viability) and adjacent segregation products in mature spermatozoa ((207 spermatozoa/18 adjacent products)/(183 spermatozoa/2 alternate products)). Therefore, as a working hypothesis, ∑p reflects the proportion of female segregation products and 1–(∑p × s) provides an estimate of the degree of germ cell death (83%). Using this approach the proportion of products consistent with 3:3 alternate segregation was estimated to be 4.7% for female heterozygotes compared with 28.3% for male carriers. Although the numbers are small, empirically embryos (Supplementary Appendix IV) from female heterozygotes have a lower proportion of normal/balanced biopsy results (4.9%, 95% CI 0.6–16.5%) compared with male carriers (19.4%, 95% CI 8.2–36.0%) (difference 296%, P=0.042).
Pachytene hexavalent diagrams illustrating the different segregation modes at meiosis I for the 64-product model. The arrows indicate the direction of separation to each pole. 1.1. 3:3 alternate segregation resulting in A,B,C and der(A),der(B),der(C). 1.2. 3:3 adjacent segregation (reflecting adjacent-1) resulting in A,B,der(C) and der(A),der(B),C; there are four other possible products from this mode. 1.3. 3:3 adjacent segregation (reflecting adjacent-2) resulting in A,der(A),C and B,der(B),der(C); there are 10 other possible products from this mode. 1.4. 4:2 segregation resulting in A,B,C,der(C) and der(A),der(B); there are 28 other possible products from this mode. 1.5. 5:1 segregation resulting in A,der(A),B,der(B),C and der(C); there are 10 other possible products from this mode. 1.6. 6:0 segregation resulting in A,der(A),B,der(B),C,der(C) and null.
To assess a translocation, the input measures are the sizes of each of the segments (the three centric segments containing the centromere and the three translocated segments, in millimetres using the 850 bphs ISCN 201319 ideogram) and a corresponding unbalanced live birth risk using single-segment estimates derived for terminal exchanges,20, 21 which are used to estimate the viability of each centric and translocated segment. Assigning imbalance thresholds (deletion and gain expressed as the proportion of the haploid autosomal length), the model calculates the overall degree of chromosomal loss (monosomy) and gain (trisomy) for each unbalanced product and identifies those with the potential to be clinically significant. To assess the performance of a potential PGD test using FISH, the input measures are the lymphocyte work-up probe efficiencies (the proportion of diploid nuclei with fewer than two signals and more than two signals) for each segment tested.22
The output measures calculated include: the natural conception live birth risk of chromosome imbalance, test accuracy, sensitivity, specificity, predictive value and likelihood of obtaining a transferable result, and the degree of risk reduction using PGD.
Results
PGD cycles and clinical outcome
Cycle and clinical outcome details are presented in Table 3. Six cycles were started, one each for Case A and Case C and two each for Case B and Case D, and all six progressed to egg collection and biopsy. There were two embryo transfers, the second cycle for Case B had one embryo transferred and the cycle for Case C had two embryos transferred; both resulted in the delivery of a healthy singleton infant with no structural malformations (not known to have been karyotyped).
Biopsied cells from a total of 37 embryos were tested on day 3 for the translocation chromosomes. Of 29 embryos from the male heterozygotes, 6 (20.7%) were diagnosed to be normal or balanced and 23 (79.3%) to be unbalanced. Of eight embryos from the female heterozygote, none was diagnosed to be normal or balanced. Confirmation of diagnosis was carried out for the embryos from the male heterozygotes but was not possible for the female carrier.
Test results and confirmation of diagnosis
Supplementary Appendix V shows the 29 male heterozygote embryo biopsy results and the confirmation of diagnosis; 29 (100%) embryos had a successful biopsy result; 6 (20.7%) embryos had a normal/balanced result and 23 (79.3%) an abnormal result.
Of the six embryos with a normal/balanced result, one could not be investigated further as it was transferred without implantation, one was discarded, and two were cryopreserved. Two normal/balanced results were confirmed by the delivery of healthy infants.
Out of 23 embryos that had abnormal results, 15 (65.2%) were confirmed to be uniformly unbalanced (true abnormal), 2 (8.7%) were found to be normal/balanced (false abnormal, 1 consistent with FISH error and 1 with FISH error or mosaicism), 4 (17.4%) had two or more unbalanced near-diploid cell lines or polyploidy (true abnormal, mosaic), 1 (4.3%) had an unbalanced complex chromosome complement (true abnormal, chaotic), and 1 (4.3%) had only fragmented nuclei (true abnormal, fragmented).
Diagnostic accuracy
Both offspring with a normal/balanced biopsy (index) result were healthy, and, although not karyotyped, presumed to have a normal (balanced) complement, and the four embryos that did not have a confirmation result (reference standard) were also initially assigned as normal for the chromosomes tested. Out of 29 index results (Supplementary Appendix VI) there were 2 (6.9%) false abnormal results and no false normal results. The test accuracy was estimated to be 93.1% (27/29) with 100% (21/21) sensitivity, and 75% (6/8) specificity. With 72.4% (21/29) prevalence, the predictive value was estimated to be 91.3% (21/23) for an abnormal test result and 100% (6/6) for a normal test result.
One transferred embryo with a normal index result did not implant and therefore could not be confirmed. Following sensitivity analysis (Supplementary Appendix VI), which assumed that the biopsy diagnosis for this embryo could be incorrect, the test accuracy was 89.7% (26/29) with 95.5% (21/22) sensitivity and 71.4% (5/7) specificity; with 75.9% (22/29) prevalence, the predictive value was 91.3% (21/23) for an abnormal test result and 83.3% (5/6) for a normal test result.
Translocation meiotic segregation products
Table 4 shows that out of 29 embryos 8 (27.6%) were consistent with 3:3 alternate segregation resulting in a normal or balanced CCR chromosome complement. Twelve (41.4%) unbalanced embryos were consistent with 3:3 adjacent segregation of the translocations, of which two (6.9%) reflected adjacent-1 and 10 (34.5%) adjacent-2 segregation. Seven (24.1%) unbalanced embryos were consistent with 4:2 nondisjunction and none was found with 5:1 or 6:0 segregation. The probable mode of meiotic segregation could not be ascertained for two (6.9%) embryos due to complex mosaicism or nucleus fragmentation. We found two embryos that were consistent with 3:3 adjacent-2 segregation following an odd number of crossovers in the interstitial segment between the centromere and the breakpoint: Case A embryo 5–6,der(6),der(6),11,der(11),16 and Case B embryo 9-1,der(1),4,14,14,14.
Assessment of three-way translocations and sensitivity analysis
Supplementary Appendix VII shows the output measures for different scenarios for male and female heterozygotes using the translocation model (Supplementary Appendix III). In addition to worked examples using cases A–D, the classes of model assessed included: testing all six segments (3CS/3TS), two centric segments and three translocated segments (2CS/3TS), three centric segments and two translocated segments (3CS/2TS), and none of the centric segments and all three translocated segments (0CS/3TS). For male and female carriers, respectively, 71.67 and 95.27% prevalence was assumed and an initial 95% efficiency per probe, which was varied in a sensitivity analysis.
For male heterozygotes the predictive values for a normal and abnormal result, respectively, were greater than 99 and 90% where at least five probes were used (scenarios 1 and 2) and 78 and 94% using only three probes for the translocated segments (scenario 3). At least 95% NPV and 85% PPV were obtained where the efficiency for each probe was at least 88.92% (scenario 5) and 90.62% (scenario 4) using five and six probes, respectively, and also for case A (scenario 6, 93–97% per probe), case B (scenario 7, 86–89% per probe), and case C (scenario 8, 95.5–97% per probe).
For female heterozygotes the predictive value of a normal result was 99% (scenario 9), 90% (scenario 10), and 25% (scenario 11) using six, five, and three probes, respectively; the predictive value of an abnormal result was >98% for all three scenarios. At least 95% NPV and 85% PPV (the test pass thresholds) were obtained where the efficiency for each probe was at least 87.3% (scenario 12) and 97.4% (scenario 13) using six and five probes, respectively; however, for case D (scenario 14, five probes, 90–97% per probe) the predictive value was only 88.3% for a normal result, indicating an 11.7% risk of misdiagnosis.
For male heterozygotes, using a PGD test with at least five probes reduced the risk of an unbalanced conception to <1% with at least a 20% chance of a transferable result per embryo tested and a ≥75% chance of transfer-testing six embryos (the median number in our experience for couples with a translocation16). Using only three probes for the translocated segments, the risk of an unbalanced translocation product after PGD was 22.37%. For female carriers, a <1% risk (with only a 3.51% chance of a transferable result and a 19.3% chance of transfer-testing six embryos) was only achieved using six probes, and the risk was 9.52% (4.05% transferable) and 75.29% (16.42% transferable) using five and three probes, respectively.
Using the models, the risk of a natural pregnancy live birth with an unbalanced translocation was estimated to be <1% for cases A and B, 5.8% for case C, and 17.9% for case D (the female heterozygote).
Discussion
This paper describes our findings following six PGD cycles for four couples with three-way translocations (three male and one female heterozygote), which resulted in a healthy singleton live birth for two couples where the male partner carried the translocation.
There have been three published studies on sperm analysis for male carriers of three-way translocations.18, 23, 24 In our study and the sperm study of Pellestor et al18 the proportions of the products were similar, and unbalanced products reflecting 3:3 adjacent-2 segregation and those consistent with 4:2 nondisjunction were much higher than the proportions consistent with 2:2 adjacent-2 and 3:1 in sperm and embryo studies of two-way reciprocal translocations.16, 25 We are aware of two other published reports that included PGD for three-way translocations. Escudero et al26 reported five cycles for a female t(5;13;16) heterozygote aged 33 years where one cell was biopsied from each embryo and tested using six probes for the translocation. A total of 22 embryos were analysed; 20 were diagnosed to be abnormal and two to be normal/balanced, one of which was transferred in the first cycle, resulting in a live birth. Lim et al27 reported one cycle for a female carrier of a t(6;10;8) aged 33 years and one cycle for a male t(5;13;8) heterozygote whose partner was aged 33 years. For both cases the strategy was to biopsy two cells from each embryo; each nucleus was tested with a different probe panel comprising a total of five probes for each translocation. Eleven embryos were analysed for the t(6;10;8) and none was diagnosed to be normal/balanced. Of seven embryos analysed for the t(5;13;8), one was diagnosed to be normal/balanced and transferred but no pregnancy resulted. Therefore, in total, including our cases, 19.4% (probably between 8.2 and 36.0%) of cleavage-stage embryos for male heterozygotes and 4.9% (probably between 0.6 and 16.5%) for female carriers had a normal/balanced result.
However, the most complete reporting of products is available from the study of Pellestor et al18 Using this study and applying a selection coefficient calculated from the relative proportion of 3:3 alternate and adjacent products, we estimated the proportion of products before selection as a working hypothesis of what might be expected for the female heterozygote. We calculated that the proportion of products consistent with 3:3 alternate segregation could be 4.7% for females compared with 28.3% for males (Table 2), which reflects the empirical observation in embryos. Although further studies are required, a differential of this order has implications for the degree of reproductive risk for both sexes and the effectiveness of PGD for female heterozygotes.
PGD for infertile CCR carriers may have the utility to improve the efficiency of assisted conception, and for fertile couples, to reduce the risk of an affected child or miscarriage due to an unbalanced product of the rearrangement. For others, the distress of pregnancy termination, which is ethically unacceptable to some couples, might be avoided. Provided that a potential FISH test has two probes that are diagnostic for potentially viable imbalance and at least one probe for five of the six translocation segments, PGD should have high accuracy and significantly reduce the risk of an unbalanced conception. It is worth noting that two of the male heterozygotes in our study had very few sperms and poor parameters, which would preclude any chance of natural pregnancy. However, the chance of obtaining at least one embryo for transfer following egg collection may be substantially less for female carriers (around one in five biopsy cycles testing on average six embryos) compared with males (around three in four biopsy cycles).
We found that genetic testing using FISH had a high degree of accuracy. Our results indicate that the diagnostic accuracy of PGD using the FISH technique for three-way translocations is similar to two-way reciprocal translocations,16 and an error rate of 6.9–10.3% is comparable to the 6–10% reported by others in PGD for chromosome rearrangements.28
A private reproductive risk assessment based on a sufficient reproductive history of the family is not possible for most CCR couples. A 20% risk of live-born offspring with an unbalanced translocation and an up to 50% risk of miscarriage have been recommended as appropriate general risk figures,21 which is largely based on the obstetric history of female heterozygotes.4, 29 Due to spermatocyte arrest, very few male three-way translocation heterozygotes are likely to conceive without assisted conception; due to small numbers caution is advised but accepting a higher incidence of 3:3 alternate products, it could be appropriate to consider a lower general risk figure of around 3% (probably between 1 and 8%, Supplementary Appendix VIII) for live birth with chromosome imbalance. However, we suggest that a risk specific for each translocation, estimated using the method we have outlined (Supplementary Appendix III), is more useful in helping a couple decide whether PGD is preferable to natural pregnancy and prenatal diagnosis.
The application of array comparative genomic hybridization and single nucleotide polymorphism microarray to PGD has the potential to offer an efficient test for complex chromosome rearrangements30 while also testing for other genome imbalance.31, 32, 33, 34 However, as with a FISH approach, comprehensive assessment (for instance using telomere probes and chromosome painting) of the rearrangement is necessary before PGD to exclude unexpected complexity that may not be detected by karyotyping alone. We suggest that a cautious approach to PGD for complex chromosome rearrangements is advisable, especially in de novo cases or where the family history information is restricted and there has been minimal characterization to provide reassurance that the rearrangement is not more complex.
In conclusion, our study shows that PGD for three-way translocations using the FISH technique with at least five probes has high accuracy and can be effective for male heterozygotes, even with very poor sperm parameters that would preclude natural pregnancy. However, there is an indication that for female carriers, the incidence of 3:3 alternate segregation could be significantly lower, to an extent where PGD may not be their best reproductive option and gamete donation or adoption may be more realistic. It is established that the profiles of segregation products for carriers of Robertsonian and reciprocal translocations show differences between female and male carriers,16, 17, 21, 35 underlining the different mechanisms and checkpoints involved in male and female meiosis. Our study reported here indicates that meiotic mechanisms in CCR carriers have similar constraints to those in other chromosome rearrangements.
References
Madan K : What is a complex chromosome rearrangement? Am J Med Genet Part A 2013; 161A: 1181–1184.
Pai GS, Thomas GH, Mahoney W, Migeon BR : Complex chromosome rearrangements. Report of a new case and literature review. Clin Genet 1980; 18: 436–444.
Kausch K, Haaf T, Köhler J, Schmid M : Complex chromosomal rearrangement in a woman with multiple miscarriages. Am J Med Genet 1988; 31: 415–420.
Madan K, Nieuwint AW, van Bever Y : Recombination in a balanced complex translocation of a mother leading to a balanced reciprocal translocation in the child. Review of 60 cases of balanced complex translocations. Hum Genet 1997; 99: 806–815.
Madan K : Balanced complex chromosome rearrangements: reproductive aspects. A review. Am J Med Genet A 2012; 158A: 947–963.
Pellestor F, Anahory T, Lefort G et al: Complex chromosomal rearrangements: origin and meiotic behavior. Hum Reprod Update 2011a; 17: 476–494.
De Gregori M, Ciccone R, Magini P et al: Cryptic deletions are a common finding in "balanced" reciprocal and complex chromosome rearrangements: a study of 59 patients. J Med Genet 2007; 44: 750–762.
Feenstra I, Hanemaaijer N, Sikkema-Raddatz B et al: Balanced into array: genome-wide array analysis in 54 patients with an apparently balanced de novo chromosome rearrangement and a meta-analysis. Eur J Hum Genet 2011; 19: 1152–1160.
Ogilvie CM, Raymond FL, Harrison RH, Scriven PN, Docherty Z : A new approach to the elucidation of complex chromosome rearrangements illustrated by a case of Rieger syndrome. J Med Genet 1998; 35: 234–237.
Saadallah N, Hulten M : A complex three breakpoint translocation involving chromosomes 2, 4, and 9 identified by meiotic investigations of a human male ascertained for subfertility. Hum Genet 1985; 71: 312–320.
Johannisson R, Löhrs U, Passarge E : Pachytene analysis in males heterozygous for a familial translocation (9;12;13) (q22; q22; q32) ascertained through a child with partial trisomy 9. Cytogenet Cell Genet 1988; 47: 160–166.
Pickering S, Polidoropoulos N, Caller J, Scriven P, Ogilvie CM, Braude P, Preimplantation Genetic Diagnosis Study Group: Strategies and outcomes of the first 100 cycles of preimplantation genetic diagnosis at the Guy's and St. Thomas' Center. Fertil Steril 2003; 79: 81–90.
Scriven PN, Kirby TL, Ogilvie CM : FISH for pre-implantation genetic diagnosis. J Vis Exp 2011; 48: e2570.
Scriven PN, Handyside AH, Ogilvie CM : Chromosome translocations: segregation modes and strategies for preimplantation genetic diagnosis. Prenat Diagn 1998; 18: 1437–1449.
Mackie OC, Scriven PN : Meiotic outcomes in reciprocal translocation carriers ascertained in 3-day human embryos. Eur J Hum Genet 2002; 10: 801–806.
Scriven PN, Flinter FA, Khalaf Y, Lashwood A, Mackie OC : Benefits and drawbacks of preimplantation genetic diagnosis (PGD) for reciprocal translocations: lessons from a prospective cohort study. Eur J Hum Genet 2013; 21: 1035–1041.
Bint SM, Ogilvie CM, Flinter FA, Khalaf Y, Scriven PN : Meiotic segregation of Robertsonian translocations ascertained in cleavage-stage embryos—implications for preimplantation genetic diagnosis. Hum Reprod 2011; 26: 1575–1584.
Pellestor F, Puechberty J, Weise A et al: Meiotic segregation of complex reciprocal translocations: direct analysis of the spermatozoa of a t(5;13;14) carrier. Fertil Steril 2011b; 95: e17–e22.
ISCN (2013): An International System for Human Cytogenetic Nomenclature, Shaffer LG, McGowan-Jordan J, Schmid M (eds), S. Karger: Basel, Switzerland, 2013.
Stengel-Rutkowski S, Stene J, Gallano P : Risk estimates in balanced parental reciprocal translocations. Monographies des Annales de Génétique 1988, Expansion Scientifique Française, Paris.
Gardner RJM, Sutherland GR, Shaffer LG : Chromosome Abnormalities and Genetic Counseling 4th Ed 2012 Oxford University Press: New York, USA.
Scriven PN, Bossuyt PM : Diagnostic accuracy: theoretical models for preimplantation genetic testing of a single nucleus using the fluorescence in situ hybridization technique. Hum Reprod 2010; 25: 2622–2628.
Cifuentes P, Navarro J, Míguez L, Egozcue J, Benet J : Sperm segregation analysis of a complex chromosome rearrangement, 2;22;11, by whole chromosome painting. Cytogenet Cell Genet 1998; 82: 204–209.
Loup V, Bernicot I, Janssens P et al: Combined FISH and PRINS sperm analysis of complex chromosome rearrangement t(1;19;13): an approach facilitating PGD. Mol Hum Reprod 2010; 16: 111–116.
Estop AM, Van Kirk V, Cieply K : Segregation analysis of four translocations, t(2;18), t(3;15), t(5;7), and t(10;12), by sperm chromosome studies and a review of the literature. Cytogenet Cell Genet 1995; 70: 80–87.
Escudero T, Estop A, Fischer J, Munne S : Preimplantation genetic diagnosis for complex chromosome rearrangements. Am J Med Genet A 2008; 146A: 1662–1669.
Lim CK, Cho JW, Kim JY, Kang IS, Shim SH, Jun JH : A healthy live birth after successful preimplantation genetic diagnosis for carriers of complex chromosome rearrangements. Fertil Steril 2008; 90: 1680–1684.
Munné S, Sandalinas M, Escudero T, Fung J, Gianaroli L, Cohen J : Outcome of preimplantation genetic diagnosis of translocations. Fertil Steril 2000; 73: 1209–1218.
Gorski JL, Kistenmacher ML, Punnett HH, Zackai EH, Emanuel BS : Reproductive risks for carriers of complex chromosome rearrangements: analysis of 25 families. Am J Med Genet 1988; 29: 247–261.
Vanneste E, Melotte C, Voet T et al: PGD for a complex chromosomal rearrangement by array comparative genomic hybridization. Hum Reprod 2011; 26: 941–949.
Alfarawati S, Fragouli E, Colls P, Wells D : First births after preimplantation genetic diagnosis of structural chromosome abnormalities using comparative genomic hybridization and microarray analysis. Hum Reprod 2011; 26: 1560–1574.
Fiorentino F, Spizzichino L, Bono S et al: PGD for reciprocal and Robertsonian translocations using array comparative genomic hybridization. Hum Reprod 2011; 26: 1925–1935.
Treff NR, Northrop LE, Kasabwala K, Su J, Levy B, Scott RT Jr. : Single nucleotide polymorphism microarray-based concurrent screening of 24-chromosome aneuploidy and unbalanced translocations in preimplantation human embryos. Fertil Steril 2011; 95: 1606–1612.
van Uum CM, Stevens SJ, Dreesen JC et al: SNP array-based copy number and genotype analyses for preimplantation genetic diagnosis of human unbalanced translocations. Eur J Hum Genet 2012; 20: 938–944.
Stene J, Stengel-Rutkowski S : Genetic risks of familial reciprocal and Robertsonian translocation carriers; In Daniel A (ed.): The Cytogenetics of mammalian autosomal rearrangements. Alan R Liss: New York, USA, 1988, pp 3–72.
Acknowledgements
We are grateful to Toby Kirby for his first-class technical expertise.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Scriven, P., Bint, S., Davies, A. et al. Meiotic outcomes of three-way translocations ascertained in cleavage-stage embryos: refinement of reproductive risks and implications for PGD. Eur J Hum Genet 22, 748–753 (2014). https://doi.org/10.1038/ejhg.2013.237
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2013.237
Keywords
This article is cited by
-
Characterization of a complex chromosomal rearrangement involving chromosomes 1, 3, and 4 in a slightly affected male with bad obstetrics history
Journal of Assisted Reproduction and Genetics (2018)
-
A novel male 2;4;14 complex chromosomal translocation with normal semen parameters but 100% embryonic aneuploidy
Journal of Assisted Reproduction and Genetics (2018)