Abstract
Copy number variants visible with the light microscope have been described as euchromatic variants (EVs) and EVs with extra G-light material at 8q21.2 have been reported only once before. We report four further patients with EVs of 8q21.2 ascertained for clinical (3) or reproductive reasons (1). Enhanced signal strength from two overlapping bacterial artificial chromosomes (BACs) and microarray analysis mapped the EV to a 284-kb interval in the reference genome. This interval consists of a sequence gap flanked by segmental duplications that contain the 12-kb components of one of the largest Variable Number Tandem Repeat arrays in the human genome. Using digital NanoString technology with a custom probe for the RNA exonuclease 1 homologue (S. cerevisiae)-like 1 (REXO1L1) gene within each 12-kb repeat, significantly enhanced diploid copy numbers of 270 and 265 were found in an EV family and a median diploid copy number of 166 copies in 216 controls. These 8q21.2 EVs are not thought to have clinical consequences as the phenotypes of the probands were inconsistent, those referred for reproductive reasons were otherwise phenotypically normal and the REXO1L1 gene has no known disease association. This EV was found in 4/3078 (1 in 770) consecutive referrals for chromosome analysis and needs to be distinguished from pathogenic imbalances of medial 8q. The REXO1L1 gene product is a marker of hepatitis C virus (HCV) infection and a possible association between REXO1L1 copy number and susceptibility to HCV infection, progression or response to treatment has not yet been excluded.
Similar content being viewed by others
INTRODUCTION
At least 1% of individuals have a copy number variation (CNV) larger than 1 Mb1 and, when copy number is high enough, CNVs become visible with the light microscope. These have been described as euchromatic variants (EVs)2 (Chromosome Anomaly Collection) and involve (1) constitutional cytogenetic amplification of known CNV regions or (2) duplication, triplication or deletion of large segmentally duplicated (SD) euchromatic tracts which may include heterochromatin and accompany pseudo-dicentric variants (Table 1).3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25 Most of the EVs are clinically innocuous but the 8p23.1 EV has been associated with a number of traits5, 6, 7, 8 and the 4p16.1 EV co-segregated with microtia.4 EVs need to be distinguished from pathological imbalances with which they may be similar or even identical under the light microscope.5, 19, 21, 24, 25
Recently, Manvelyan et al9 reported three patients of Middle European and Japanese descent with a microscopically visible CNV of 8q21.2. This CNV was characterised by a 60% increase in the signal strength from a single bacterial artificial chromosome (BAC) but an extra copy of a single BAC of ∼123 kb would not normally account for the microscopic visibility of this 8q21.2 EV. Here, we have confirmed the existence of this 8q21.2 EV in four new patients from Canada (Table 2) and set out to refine the location and extent of this EV. We have used digital NanoString technology to quantify the copy number of the variable number tandem repeat (VNTR) array within the EV and find that expansion of this 8q21.2 VNTR is common and most likely to account for the microscopic visibility of the 8q21.2 EV. We estimate the prevalence of the 8q21.2 EV among postnatal referrals for chromosome analysis and suggest that CNV of the RNA exonuclease 1 homologue (S. cerevisiae)-like 1 (REXO1L1) gene cluster within the VNTR may be related to susceptibility to hepatitis C infection.
METHODS
Chromosomes were prepared and analysed using standard procedures on a total of 3078 consecutive clinical referrals for peripheral blood analysis over a 21 month period. Single- or dual-colour fluorescence in situ hybridisation (FISH) was performed with BACs from 8q21.2, a centromere probe for chromosome 8 (D8Z2) and, for Patient 4, a centromere probe for chromosomes 9 (D9Z3) with a centromere probe for chromosome 11 as a control (Cytocell, Cambridge, UK). DNA was extracted from peripheral blood using Puregene DNA Isolation Kits (Gentra, Minneapolis, MN, USA) and used for microarray analysis with Affymetrix Cytogenetics 2.7M (Patient 1) or Affymetrix Cytoscan HD (Patient 4) microarrays according to the manufacturer’s instructions (Affymetrix, Santa Clara, CA, USA). The Affymetrix CEL data files were converted into CYCHP files using the annotation files na30.2 (Patient 1; 2.7M array) or na32.1 (Patient 4; Cytoscan HD array) and the microarray data were analysed using the Affymetrix Chromosome Analysis Suite (ChAS) Software v1.1 (Patient 1; 2.7M array) or v1.2 (Patient 4; Cytoscan HD array) (Affymetrix). The reference model file was provided by Affymetrix. Quality control parameters were SNPQC ≥1.1 and MAPD ≤0.27 (Patient 1; 2.7M array) or SNPQC ≥15 and MAPD ≤0.25 (Patient 4; Cytoscan HD array). Base pair coordinates correspond or have been converted to release GRCh37 (hg19). DNA for NanoString analysis was extracted using standard methods and used for genomic copy number analysis with a custom NanoString code set comprising 10 normalisation probes targeting invariant autosomal loci, and 27 custom probes targeting highly CNV sites including one targeting the REXO1L1 gene cluster (probe sequence GTTTCCCCTACAAGCGCTCCCTCAGGAATCTCGCGGCCGACTACCTGGCACAGATCATCCAGGACAGCCA). Patient DNA samples and 216 anonymous controls of self-reported European ancestry were processed using the NanoString nCounter Analysis System (NanoString Technologies, Seattle, WA, USA)26 according to the manufacturer’s instructions. The ethnicity of patients and/or parents was based on their self-reported national origins.
Clinical details
Patient 1 was a woman of 33 referred for recurrent miscarriages, Patient 2 a woman of 26 with secondary amenorrhoea referred to exclude a possible diagnosis of Turner’s syndrome, Patient 3 a boy of 3 with autism and Patient 4 a newborn girl with a large ventricular septal defect and dysmorphic features (mild hypotelorism, abnormal palpebral fissures and a smooth, thin upper lip). Her father was phenotypically normal and healthy.
RESULTS
Our results and those of Manvelyan et al9 are summarised in Table 2.
Karyotyping
Additional G-light material was found at band 8q21.2 in all four index patients (Figures 1a–d) among 3078 consecutive clinical referrals for peripheral blood analysis over a 21-month period. The additional material could have been the result of duplication, insertion, paracentric inversion or conformational alteration. However, the banding pattern was not consistent with an inversion and the extra material appeared to be cytogenetically identical to the microscopically visible CNVs described by Manvelyan et al.9 The same variant was found in the father of Patient 4 (Figure 1e) and, following microarray analysis of Patient 4 (see below), mosaic trisomy 9 was confirmed in 2/200 G-banded metaphases (data not shown). Other parental samples were requested but not received.
Idiogram of a normal chromosome 8 and partial G-banded karyotypes of the EV chromosomes 8 with the variant chromosome on the right in each case and the additional G-light material indicated by the black arrow: (a) Patient 1; (b) Patient 2; (c) Patient 3; (d) Patient 4 and (e) the father of Patient 4.
FISH
Using a panel of five BACs from medial 8q, Manvelyan et al9 found that only BAC RP11-96G1 gave signals that were 60% larger on the EV chromosome than on the normal chromosome. We noted that RP11-96G1 overlaps with RP11-90G23 and that RP11-90G23 contains segmental duplications (SDs) of 8q21.2 that might mediate amplification of the sequence between them, represented as gap in the human genome assembly (hg19). Enhanced signals on one chromosome 8 were consistently found using single- or dual-colour FISH with both BACs in all four new patients (Figures 2a–d) but the increase in size was more consistent with duplication than with amplification (Figures 2a–d). However, these larger signals could not be separated in prometaphase (Figure 2c) or interphase cells (Figure 2a). Following microarray analysis (see below), mosaic trisomy 9 was found in Patient 4 and confirmed in 14/200 interphase nuclei (data not shown).
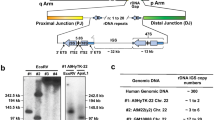
(a–e) FISH and microarray results: (a, b) single-colour FISH with BACs RP11-90G23 (green) and BAC RP11-96G1 (red) in metaphases and an interphase nucleus from Patient 1 with the variant chromosome indicated by the large arrows; (c) dual-colour FISH with BACs RP11-90G23 (green) and RP11-96G1 (red) in an interphase nucleus from Patient 1 showing the colocalisation of the combined signals (yellow); (d) FISH with BAC RP11-96G1 (red) in a prometaphase from Patient 1 showing the lack of separation of the signals on the variant chromosome; (e) idiogram of chromosome 8 showing the location of the CNV region in distal 8q21.2; (f) Affymetrix Cytoscan HD array analysis of base pairs 86 460 508–87 259 456 in Patient 4 using the Affymetrix ChAS Software. The normal copy number results cluster along the horizontal weighted log2 ratio=0 line and flank the variant 284 kb region between base pairs 86 553 129 and 86 836 909 (hg19). The horizontal lines above and below represent log2 ratios of +1 and −1, respectively. (g) A Portable Document Format (PDF) screen shot of the variant and flanking regions of 8q21.2 from the UCSC browser (GRCh37/hg19). The FISH clones are in green with RP11-90G23 manually annotated from BAC end sequence data (base pairs 86 570 255–86 817 480). The flanking SDs are represented by black double-headed arrows between black vertical lines and have been labelled REPP (for REPeat Proximal) and REPD (for REPeat Distal). The sequence gap between the SDs is represented by the black bar labelled GAP. The RefSeq genes are in blue with the tandem REXO1L1 gene array highlighted by the red arrows underneath. The region contains no non-coding sequences. The DGV tracks show gains (blue), losses (red) and inversions (brown). The UCSC SD track is mostly in terracotta.
Microarray
Affymetrix Cytogenetics Whole-Genome 2.7M Array analysis in Patient 1 and Cytoscan HD array analysis in Patient 4 were normal except that evidence of low-level trisomy 9 mosaicism was detected in Patient 4 and subsequently confirmed using FISH and karyotyping (see above) (data not shown). DNA for microarray testing was requested from Patient 3 but not received, and not thought to be warranted in Patient 2 on clinical grounds. The normal array results exclude a duplication of the great majority of chromosome 8 covered by the arrays or an unbalanced insertion into 8q from elsewhere. The array results also localise the variant region to a 284-kb interval in distal 8q21.2 (Figure 2e) between base pairs 86 553 129 and 86 836 909 (hg19) in Patient 4 (Figure 2f). The corresponding 174 kb interval between base pairs 86 738 256 and 86 912 598 (hg18) in Patient 1 converts to an almost identical 292 kb between base pairs 86 551 004 and 86 843 310 in the current build of the human genome (hg19). This common region in Patients 1 and 4 contains the sequence gap and flanking SDs REPP and REPD in the reference genome (Figure 2g).
The karyotype of the index Patients 2 and 3 was 46,XX or XY,var(8)(q21.2q21.2).ish var(8)(RP11-90G23enh,RP11-96G1enh), the karyotype of index Patient 1 was 46,XX,var(8)(q21.2q21.2).ish var(8)(RP11-90G23enh,RP11-96G1enh).arr(1-22)x2 and that of Patient 4 was mos 46,XX,var(8)(q21.2q21.2)[198]/47,XX,var(8)(q21.2q21.2),+9[2].ish var(8)(q21.2q21.2)(RP11-90G23enh,RP11-96G1enh).nuc ish(D9Z3 × 3,D11Z1 × 2)[14/200].arr(9)(1–141 213 431)x2∼3.
Molecular genetics
Pulse-field electrophoresis (PFGE) had previously demonstrated that the 8q21.2 SDs contain elements of a tandem VNTR array consisting of 50–150 copies of a 12-kb repeat that spanned 0.6–1.7 Mb in two pedigrees.27 We used digital NanoString technology with a custom probe for the REXO1L1 gene cluster to determine total (diploid) copy number of the 12 kb repeat. There was a close linear correlation between total copy number measured using PFGE Southern blot and the NanoString nCounter Analysis System (R2=0.98) (Figure 3a). Total copy numbers of 265 and 270 were found in Patient 4 and her father, respectively, compared with a median of 166 and an average of 173 in 216 normal controls (Figure 3b). Copy numbers ranged from 97 to 277 in the controls with a positive skew of 0.7 (Figure 3b) and, by comparison, the copy numbers in the EV family were greater than the 99th centile and significantly higher using the Mann–Whitney Rank Sum Test (P=0.018).
(a) Comparison of the total (diploid) copy number of the REXO1L1 gene cluster measured by PFGE Southern blot and the NanoString nCounter Analysis System showing the close linear correlation; (b) distribution of the frequency of REXO1L1 copy numbers (y axis) binned into intervals of 10 copies from 90 to 280 (x axis) in 216 controls. The median copy number of 166 is indicated by the dotted line labelled M, the mean value of 173 by the dashed line labelled A and the copy numbers of 265 and 270 in Patient 4 and her father represented by the black stars.
DISCUSSION
Our results map the 8q21.2 EVs to the sequence gap and flanking SDs in distal 8q21.2 that are clearly larger than 284 kb in the reference genome (Figures 2e and g) and most likely to underlie this EV. Compared with the other 16 EVs in Table 1, the 8q21.2 EVs have by far the smallest amplicon, the highest copy number and, in common with the 4p16.1 and 8p23.1 EVs, are flanked by SDs of the kind that underlie the majority of other EVs.
The NanoString analysis indicates that an average of 173 diploid copies of the REXO1L1 gene cluster and 12 kb repeat are present within the 8q21.2 VNTR in our 216 controls. This implies that an average chromosome 8 contains 87 copies spanning over 1 Mb or almost half of band 8q21.2 which is a predicted 2.3 Mb in the UCSC browser28 or an estimated 2.1 Mb (assuming that 8q21.2 is 2% of 8q29 and that 8q is 105 Mb30). If accompanied by an average chromosome 8, then the 265 and 270 copies in Patient 4 and her father imply EV chromosomes with∼180 copies spanning over 2.1 Mb. Thus, given the ∼2:1 ratio of the enhanced FISH signals (Figures 2a–d),9 the simplest interpretation of our results is that an average VNTR array of∼90 copies is duplicated to give an EV chromosome of∼180 copies, a diploid total of 270 copies and a differential length of at least 1 Mb (Figure 2g). However, the EV FISH signals could not be resolved into separate blocks (Figures 2a and c) and there is a relatively even distribution of copy numbers from a minimum of 97 to a maximum of 277 in the 216 controls (Figure 3b). In addition, the copy numbers in EV carriers are significantly higher than the great majority of controls and above the 99th centile of the control range. We therefore propose that expansion of the VNTR array is most likely to underlie the 8q21.2 EVs, and that their microscopic visibility reflects the combination of an expanded VNTR array on the EV chromosome with an average or smaller than average VNTR array on the homologous chromosome 8. An analogous situation was found for the beta defensin repeat underlying the 8p23.1 EV: this was detected in rare instances of high diploid copy numbers unequally split between homologous chromosomes 8;31 only subsequently did the frequency of similar diploid copy numbers in control populations imply the presence of individuals with above average copy numbers on both homologous chromosomes.5 Interphase FISH has been used to determine relative haploid copy numbers but becomes impractical when amplicon size is small and copy number very high.32 Further investigations will be needed of EV and non-EV carriers in larger families using quantitative allele-specific copy number techniques33 to confirm or exclude our proposal.
In the meantime, the microscopically visible CNV of 8q21.2 is not believed to be population specific now that it has been described in seven index patients of Middle European (3), Japanese (1), Palestinian (1), Caucasian (1) and unspecified Canadian (1) descent (Table 2). These seven were ascertained with congenital anomalies, autism, secondary amenorrhoea, infertility, miscarriages and a family history of intellectual difficulties and cystic fibrosis (Table 2). However, this EV is unlikely to be a cause of congenital anomalies or autism because: (1) there was no consistency in the indications in Patients 2, 3 and 4, and the congenital anomalies in Patient 4 were most likely explained by her trisomy 9 mosaicism; (2) the three patients referred for reproductive reasons were otherwise normal (Patient 1; Manvelyan et al,9 Patients 1 and 2); (3) the 12-kb VNTR repeat array contains only the REXO1L1 gene cluster that has no known pathology to date; (4) the variant region overlaps RP11-96G1 which is a common benign CNV34 and corresponds with locus 4603a in the Database of Genome Variants (DGVs).35 A possible association with infertility and/or miscarriage9 would need testing in much larger cohorts and is more likely to reflect bias of ascertainment.36
This EV was found in 4/3078 (1 in 770) consecutive referrals over a 21-month period which suggests that this cytogenetically visible variant is not uncommon and may be misinterpreted as a pathogenic imbalance. This frequency could be an underestimate as this EV is usually discernible only in high quality chromosome preparations of at least 550 band level resolution and may have gone undetected in lower resolution chromosomes or where band 8q21.2 is unclear. In addition, Whitby et al34 found CNVs of RP11-96G1 in 142/1275 (1 in 9) clinically affected patients, which suggests that ∼1% (9/770) of these CNVs may be microscopically visible. The DGV also indicates that the background frequency of CNVs across the 8q21.2 EV region is high with gains and losses found in 10/49 patients and controls35 and 9/90 Yoruban individuals.37 Gains were also found in 26/30 HapMap control samples from four populations, including Yorubans, European-Americans, Japanese and Chinese individuals.38 In addition, inversions between the 8q21.2 SDs have been detected in individuals of Yoruban and non-African descent.39 Interestingly, a diploid REXO1L2P median copy number of 172, almost identical to the 173 in our controls, was found using sequencing in 159 ethnically diverse subjects from the 1000 Genomes Project,32 and the Database of Genomic Structural Variation records 272 copies, within the EV range, of a 7-kb segment including the REXO1L1 gene in a Southern Kalahari hunter-gatherer (KB1).40
RP11-96G1 at 8q21.2 is the third most common benign CNV identified by Whitby et al34 using BAC array CGH and, like the second most common (RP11-122N11 at 8p23.1), becomes microscopically visible at high copy number,31 contains tandemly repeated DNA families,27 predisposes to polymorphic inversions41 and has been associated with neocentromere formation.42 The 8q21.2 VNTR is one of the nine autosomal tandem arrays >2 kb that contain a single gene and show CNV.27 Three of these nine have been associated with one or more traits including (1) higher copy number of the salivary amylase gene (AMY1) at 1q21.1 with high starch diets;43 (2) increasing copy number of the 8p23.1 beta defensin genes with higher predisposition to psoriasis,5, 6 higher human immunodeficiency virus (HIV) load7 and, possibly, resistance to influenza virus infection;8 and (3) lower copy number of the CCL3L1 gene at 17q21.1 with weak protection against malarial anaemia44 and susceptibility to chronic hepatitis C virus (HCV) infection.45 It is therefore interesting that immunological reaction against the human REXO1L1 gene product (GOR) is a marker of HCV infection including occult HCV in which antibody assays against the HCV are negative.46 This may simply be a reflection of the partial homology between the GOR epitopes and the HCV-encoded core protein sequence,46 but it is also conceivable that REXO1L1 copy number affects susceptibility to HCV infection, progression or response to treatment.47 A critical region including REXO1L1 and a number of candidate genes is amplified in a variety of cancers48 but overexpression of REXO1L1 in cancer has apparently been recorded only once.49
In conclusion, we have confirmed the existence of the 8q21.2 EV and mapped the variation to a region containing the 8q21.2 VNTR arrays with multiple copies of a 12-kb repeat and the REXO1L1 gene cluster. We propose that expansion of this VNTR explains the microscopic visibility of the 8q21.2 EVs, which need to be distinguished from pathogenic imbalances especially at prenatal diagnosis. Light microscopically visible imbalances with normal array results may be regarded as a general indicator of the possibility of novel EVs in other parts of the genome.
References
Itsara A, Cooper GM, Baker C et al: Population analysis of large copy number variants and hotspots of human genetic disease. Am J Hum Genet 2009; 84: 148–161.
Barber JCK : Directly transmitted unbalanced chromosome abnormalities and euchromatic variants. J Med Genet 2005; 42: 609–629.
Balikova I, Martens K, Melotte C et al: Autosomal-dominant microtia linked to five tandem copies of a copy-number-variable region at chromosome 4p16. Am J Hum Genet 2008; 82: 181–187.
Harada N, Takano J, Kondoh T et al: Duplication of 8p23.2: a benign cytogenetic variant? Am J Med Genet 2002; 111: 285–288.
Hollox EJ, Huffmeier U, Zeeuwen PL et al: Psoriasis is associated with increased beta-defensin genomic copy number. Nat Genet 2008; 40: 23–25.
Stuart PE, Hüffmeier U, Nair RP et al: Association of β-defensin copy number and psoriasis in three cohorts of European origin. J Invest Dermatol 2012; 132: 2407–2413.
Hardwick RJ, Amogne W, Mugusi S et al: β-defensin genomic copy number is associated with HIV load and immune reconstitution in sub-saharan Africans. J Infect Dis 2012; 206: 1012–1019.
Hardwick RJ, Machado LR, Zuccherato LW et al: A worldwide analysis of beta-defensin copy number variation suggests recent selection of a high-expressing DEFB103 gene copy in East Asia. Hum Mutat 2011; 32: 743–750.
Manvelyan M, Cremer FW, Lance J et al: New cytogenetically visible copy number variant in region 8q21.2. Mol Cytogenet 2011; 4: 1.
Lecce R, Murdolo M, Gelli G et al: The euchromatic 9p+ polymorphism is a locus-specific amplification caused by repeated copies of a small DNA segment mapping within 9p12. Hum Genet 2006; 118: 760–766.
Di Giacomo MC, Cesarano C, Bukvic N et al: Duplication of 9 p11.2-p13.1: a benign cytogenetic variant. Prenat Diagn 2004; 24: 619–622.
Kosyakova N, Grigorian A, Liehr T et al: Heteromorphic variants of chromosome 9. Mol Cytogenet 2013; 6: 14.
Wojiski SA, Rhodes CH, Brodhurst CA et al: The G positive band of the rare euchromatic 9qh variant is derived from 9p12. Appl Cytogenet 1997; 23: 125–129.
Willatt LR, Barber JCK, Clarkson A et al: Novel deletion variants of 9q13-q21.12 and classical euchromatic variants of 9q12/qh involve deletion, duplication and triplication of large tracts of segmentally duplicated pericentromeric euchromatin. Eur J Hum Genet 2007; 15: 45–52.
Joseph-George AM, He Y, Marshall CR et al: Euchromatic 9q13-q21 duplication variants are tandem segmental amplifications of sequence reciprocal to 9q13-q21 deletions. J Med Genet 2011; 48: 317–322.
Barber JC, Rodrigues R, Maloney VK et al: Another family with a euchromatic duplication variant of 9q13-q21.1 derived from segmentally duplicated pericentromeric euchromatin. Cytogenet Genome Res 2013; 141: 64–69.
Starke H, Seidel J, Henn W et al: Homologous sequences at human chromosome 9 bands p12 and q13-21.1 are involved in different patterns of pericentric rearrangements. Eur J Hum Genet 2002; 10: 790–800.
Goumy C, Mihaescu M, Tchirkov A et al: An unusual familial chromosome 9 ‘variant’ with variable phenotype: characterization by CGH analysis. Morphologie 2005; 89: 71–75.
Barber JC, Cross IE, Douglas F et al: Neurofibromatosis pseudogene amplification underlies euchromatic cytogenetic duplications and triplications of proximal 15q. Hum Genet 1998; 103: 600–607.
Fantes JA, Mewborn SK, Lese CM et al: Organisation of the pericentromeric region of chromosome 15: at least four partial gene copies are amplified in patients with a proximal duplication of 15q. J Med Genet 2002; 39: 170–177.
Carelle-Calmels N, Girard-Lemaire F, Guérin E et al: Proximal 15q familial euchromatic variant and PWS/AS critical region duplication in the same patient: a cytogenetic pitfall. Eur J Med Genet 2008; 51: 547–557.
Barber JC, Reed CJ, Dahoun SP, Joyce CA : Amplification of a pseudogene cassette underlies euchromatic variation of 16p at the cytogenetic level. Hum Genet 1999; 104: 211–218.
López Pajares I, Villa O, Salido M et al: Euchromatic variant 16p+. Implications in prenatal diagnosis. Prenat Diagn 2006; 26: 535–538.
Barber JCK, Hall V, Maloney VK et al: 16p11.2-p21.2 duplication syndrome; a genomic condition differentiated from euchromatic variation of 16p11.2. Eur J Hum Genet 2013; 21: 182–189.
Barber JCK, Brasch-Andersen C, Maloney VK et al: A novel pseudo-dicentric variant of 16p11.2-q11.2 contains euchromatin from 16p11.2-p11.1 and resembles pathogenic duplications of proximal 16q. Cytogenet Genome Res 2013; 39: 59–64.
Geiss GK, Bumgarner RE, Birditt B et al: Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol 2008; 26: 317–325.
Warburton PE, Hasson D, Guillem F et al: Analysis of the largest tandemly repeated DNA families in the human genome. BMC Genomics 2008; 9: 533.
Furey TS, Haussler D : Integration of the cytogenetic map with the draft human genome sequence. Hum Mol Genet 2003; 12: 1037–1044.
Francke U : Digitized and differentially shaded human chromosome ideograms for genomic applications. Cytogenet Cell Genet 1994; 65: 206–218.
Morton NE : Parameters of the human genome. Proc Natl Acad Sci USA 1991; 88: 7474–7476.
Hollox EJ, Armour JA, Barber JC : Extensive normal copy number variation of a beta-defensin antimicrobial-gene cluster. Am J Hum Genet 2003; 73: 591–600.
Sudmant PH, Kitzman JO, Antonacci F et al: Diversity of human copy number variation and multicopy genes. Science 2010; 330: 641–646.
Alkan C, Kidd JM, Marques-Bonet T et al: Personalized copy number and segmental duplication maps using next-generation sequencing. Nat Genet 2009; 41: 1061–1067.
Whitby H, Tsalenko A, Aston E et al: Benign copy number changes in clinical cytogenetic diagnostics by array CGH. Cytogenet Genome Res 2008; 123: 94–101.
Iafrate AJ, Feuk L, Rivera MN et al: Detection of large-scale variation in the human genome. Nat Genet 2004; 36: 949–951.
Barber JCK : Reply to Dundar et al: Why a causal association between classical euchromatic variants of chromosome 9 and multiple miscarriages is unlikely. Genet Couns 2010; 21: 469–471.
Matsuzaki H, Wang PH, Hu J et al: High resolution discovery and confirmation of copy number variants in 90 Yoruba Nigerians. Genome Biol 2009; 10: R125.
Perry GH, Ben-Dor A, Tsalenko A et al: The fine-scale and complex architecture of human copy-number variation. Am J Hum Genet 2008; 82: 685–695.
Kidd JM, Cooper GM, Donahue WF et al: Mapping and sequencing of structural variation from eight human genomes. Nature 2008; 453: 56–64.
Schuster SC, Miller W, Ratan A et al: Complete Khoisan and Bantu genomes from southern Africa. Nature 2010; 463: 943–947.
Giglio S, Broman KW, Matsumoto N et al: Olfactory receptor-gene clusters, genomic-inversion polymorphisms, and common chromosome rearrangements. Am J Hum Genet 2001; 68: 874–883.
Hasson D, Alonso A, Cheung F et al: Formation of novel CENP-A domains on tandem repetitive DNA and across chromosome breakpoints on human chromosome 8q21 neocentromeres. Chromosoma 2011; 120: 621–632.
Perry GH, Dominy NJ, Claw KG et al: Diet and the evolution of human amylase gene copy number variation. Nat Genet 2007; 39: 1256–1260.
Carpenter D, Färnert A, Rooth I et al: CCL3L1 copy number and susceptibility to malaria. Infect Genet Evol 2012; 12: 1147–1154.
Grünhage F, Nattermann J, Gressner OA et al: Lower copy numbers of the chemokine CCL3L1 gene in patients with chronic hepatitis C. J Hepatol 2010; 52: 153–159.
Quiroga JA, Castillo I, Bartolome J, Carren V : Serum immunoglobulin G antibodies to the GOR autoepitope are present in patients with occult hepatitis C virus (HCV) infection despite lack of HCV-specific antibodies. Clin Vaccine Immunol 2007; 14: 1302–1306.
Schaefer EA, Chung RT : The impact of human gene polymorphisms on HCV infection and disease outcome. Semin Liver Dis 2011; 31: 375–386.
Byrne JA, Chen Y, Martin La Rotta N, Peters GB : Challenges in identifying candidate amplification targets in human cancers: chromosome 8q21 as a case study. Genes Cancer 2012; 3: 87–101.
Ramakrishna M, Williams LH, Boyle SE et al: Identification of candidate growth promoting genes in ovarian cancer through integrated copy number and expression analysis. PLoS One 2010; 5: e9983.
Acknowledgements
This work was supported by NIH grant UL1RR029887 to Mount Sinai School of Medicine, NIH grants 1R01DA033660 and 1R01HG006696 to AJS, and grant 2012ALZNIRG69983 from the Alzheimer’s Association to AJS. We would like to thank all the families concerned, Monika Sharma of the University Health Network in Toronto, Canada, for her technical expertise and Viv K Maloney for the idiogram of chromosome 8.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Tyson, C., Sharp, A., Hrynchak, M. et al. Expansion of a 12-kb VNTR containing the REXO1L1 gene cluster underlies the microscopically visible euchromatic variant of 8q21.2. Eur J Hum Genet 22, 458–463 (2014). https://doi.org/10.1038/ejhg.2013.185
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2013.185
Keywords
This article is cited by
-
The structure, function and evolution of a complete human chromosome 8
Nature (2021)
-
Copy number variation of the REXO1L1 gene cluster; euchromatic deletion variant or susceptibility factor?
European Journal of Human Genetics (2017)
-
Automated, high-throughput derivation, characterization and differentiation of induced pluripotent stem cells
Nature Methods (2015)