Abstract
Three human chromosome loci (1q43, 10p12.31, and 12q21.31) were recently associated with the susceptibility to primary open-angle glaucoma (POAG) in a Japanese population; however, this was not replicated in three subsequent studies using South Indian, Afro-Caribbean, and Chinese populations. To identify genetic markers that are robustly associated across ethnic populations, numerous markers in addition to the six in the three reported loci were examined in this study. A total of 31 single-nucleotide polymorphism (SNP) markers were genotyped for 1115 Korean participants, and many neighboring SNPs were imputed using the Korean HapMap Project genotype data. Each SNP was statistically tested for association with POAG susceptibility by comparisons among 211 POAG patients with 904 unaffected controls. A strong and statistically significant association was found with a previously unreported SNP, rs7098387 (odds ratio, OR=2.0 (1.4–3.0), P=0.00038) at the 10p12.31 locus (where 11 SNPs were typed and 38 imputed) in contrast to the reported rs7081455, which was too poorly correlated with newly associated rs7098387 (r2=0.003, D′=0.40) to show association. Additionally, a modest association was observed with the reported rs693421 (OR=1.4 (1.1–1.7), P=0.0082) and several other SNPs located within and around ZP4 at the 1q43 locus (10 SNPs typed and 14 imputed). However, no association was observed with the reported rs7961953 SNP or any other SNPs at the 12q21.31 locus, upstream of TMTC2 (10 SNPs typed and 29 imputed). Accordingly, POAG susceptibility association was replicated using rs7098387 (C) rather than rs7081455 (T) at the 10p12.31 locus and additionally with rs693421 (T) at the 1q43 locus.
Similar content being viewed by others
Introduction
Glaucoma is a chronic, degenerative optic neuropathy that causes damage to the optic nerve and progresses to blindness.1 The disease involves the loss of retinal ganglion cells, leading to degeneration of the optic nerve and corresponding defects of the visual field. Elevated intraocular pressure is the strongest known risk factor for developing glaucoma and a main target of treatment for the prevention of progressive visual field loss. Although elevated pressure is clearly a risk factor, optic neuropathy can occur with normal intraocular pressure. Primary open-angle glaucoma (POAG) is the most common type of glaucoma and may or may not be accompanied by elevated intraocular pressure.2, 3, 4
POAG is known to have a genetic component. According to several population-based studies, first-degree relatives of POAG patients have a two- to fourfold increased relative risk of developing POAG when compared with the general population.5, 6, 7 The Glaucoma Inheritance Study in Tasmania revealed that POAG is familial for ∼60% of patients and that the disease is more severe in patients with a family history than those without it.8 Further, 21 POAG susceptibility loci have been identified by linkage studies, including 14 assigned as GLC1A–GLC1N and 10 that remain unassigned.1, 9, 10, 11, 12 However, only three underlying genes have been identified at these loci, MYOC in GLC1A,13 OPTN in GLC1E,14 and WDR36 in GLC1G;15 these genes account for <5% of glaucoma cases.
Recently, several POAG susceptibility loci were identified by genome-wide association (GWA) studies16, 17, 18, 19, 20 and were replicated in multiple ethnicities: 7q31.2 between CAV1 and CAV2,16, 21 9p21.3 with CDKN2B-AS1,19, 20, 22, 23, 24, 25, 26 10q21.3 with ATOH7,17, 18, 22, 27 and 14q23.1 between SIX1 and SIX6.17, 20, 22, 28 One of the GWA studies demonstrated the association of POAG with six SNPs that flanked the ZP4, PLXDC2, and TMTC2 genes located at chromosome loci 1q43, 10p12.31, and 12q21.31, respectively, in a Japanese population;29 however, these associations were not replicated in a South Indian,30 Afro-Caribbean,24 or Chinese31 population. These conflicting results can be ascribed to factors such as a small sample size with inadequate statistical power, different phenotype definitions, population-specific linkage disequilibrium (LD), effect-size bias, or population stratification.32
Association signals detected by GWA studies depend on how strongly the markers correlate with an unknown functional variation(s). Therefore, population differences in LD between an untyped, causal variation and typed markers can undermine the effect of the causal variation. Markers other than the reported ones in the association regions could be more strongly correlated with functional variations, and thereby offer greater statistical power to detect an association. A hybrid design involving exact testing of the reported markers plus fine mapping of the same region using additional markers is a good strategy for the replication and validation of the GWA study results.33, 34, 35
In this study, we investigated not only the reported SNPs but also additional SNPs that were chosen based on LD with the reported SNPs located at the loci identified from the Japanese GWA study.29 The expansive coverage led to the identification of common SNPs associated with glaucoma susceptibility in both Korean and Japanese populations.
Materials and methods
Study participants
With the approval of the Institutional Review Board of Chungnam National University Hospital and following the tenets of the Declaration of Helsinki, 1115 Korean participants were recruited at the hospital and provided written informed consent, as previously described.36 All POAG patients were diagnosed by a glaucoma specialist (C.-S.K.) at the glaucoma clinic of this tertiary referral hospital. The inclusion criteria for POAG were as follows: (1) characteristic glaucomatous defects in the optic nerve head, (2) characteristic visual field defects, as tested using the Humphrey Field Analyzer SITA standard 30-2 or 24-2 program (HFA II 720i; Carl Zeiss Meditec, Dublin, CA, USA), (3) defect in the retinal nerve fiber layer analysis using optical coherence tomography (Stratus OCT; Carl Zeiss Meditec), and (4) normal open angle by gonioscopy or deep central and peripheral anterior chamber angle width (>1/4 corneal thickness) by the van Herick method.
The POAG cases were divided into two subgroups, high-tension (intraocular pressure >21 mm Hg) and normal-tension glaucoma (intraocular pressure ≤21 mm Hg), according to the highest pressure ever recorded. The intraocular pressure was measured using a Goldmann applanation tonometer, and an average of two measurements was recorded for each visit. Patients with non-POAG types, such as exfoliative, pigmentary, steroid-induced, and primary angle-closure glaucoma, or those with uveitis were not included in the case group.
The 904 controls were free of any eye disease, and unaffected subjects having intraocular pressure higher than 21 mm Hg, cup-to-disc ratio larger than 0.7, glaucoma family history, or steroid treatment were not included in the control group. The demographic and clinical details of the study subjects are summarized in Table 1. The male/female ratio (1.6) was high in the case of this recruitment, as in the general POAG prevalence (gender ratio of 1.4) of Koreans,37 but there was no attempt to match with control recruitment (gender ratio of 1.0), which would have necessitated an adjustment for gender in the susceptibility association tests.
SNP selection and genotyping
A total of six SNPs (rs540782, rs547984, rs693421, and rs2499601 located at chromosome locus 1q43, rs7081455 at 10p12.31, and rs7961953 at 12q21.31) were reported to be associated with POAG susceptibility in a previous GWA study.29 These reported SNPs were individually in intermediate-to-strong LD with a total of 112 SNPs at D′≥0.5, with r2≥0.1, or at D′=1, with r2<0.1, in the Japanese population (JPT) of the International HapMap Project (http://hapmap.ncbi.nlm.nih.gov; Phase 2+3, release 27). These neighboring SNPs were tagged by 25 additional SNPs, at r2≥0.7, in the Korean population of the Korean HapMap Project (www.khapmap.org). The tag selection and LD calculations were performed using the HaploView 4.2 (Broad Institute, Cambridge, MA, USA), and the GenBank accession numbers of reference sequences for human 1q43, 10p12.31, and 12q21.31 loci were NT_167186.1, NT_008705, and NT_029419.12, respectively.
Genomic DNA extracted from peripheral blood leukocytes of all subjects were genotyped using the MassARRAY iPLEX Gold assay (Sequenom, San Diego, CA, USA), according to the manufacturer’s instructions, with approval by KAIST Institutional Review Board. The genotyping accuracy was 100%, as validated in blind tests of 16 samples for the six reported SNPs using the MassARRAY hME assay (Sequenom), as previously described.38 Amplification and extension primers were designed using the MassARRAY SpectroDESIGNER 3.1. The call rate per SNP ranged from 94% to 100%, and the genotypes of the control subjects were in Hardy–Weinberg equilibrium (P≥0.05/31=0.0016).
Imputation and association tests
For the association analysis, the genotype distribution between the POAG patients and controls was compared using logistic regression, with adjustments for age and gender according to the co-dominant genetic models. Association was considered statistically significant when P ≤α and marginal when α<P≤0.05, with a significance level of Bonferroni correction for multiple testing of α=0.05/31=0.0016, as 31 typed SNPs were tested. Statistical powers were calculated using the odd ratios reported in the GWA study,29 with the sample size of the present study (type II error probability of β=0.20), using the PS v.3.0 software (http://biostat.mc.vanderbilt.edu/twiki/bin/view/Main/PowerSampleSize). All the statistical tests were performed using the SPSS v.18.0 (IBM SPSS, Armonk, NY, USA) or PLINK v.1.07 package (Broad Institute, Cambridge, MA, USA).
Typing data for the 31 SNPs were used to impute the genotypes of 699 additional SNPs within a 200-kb region centered in each locus for all subjects according to the Korean HapMap Project genotype data using the MaCH 1.0.16 software (University of Michigan, Ann Arbor, MI, USA), as previously described36 (using the standard imputation approach with the - - geno command line option). Because the 25 typed tag SNPs tagged only 112 SNPs that were in intermediate-to-strong LD with the six reported SNPs, as described above, not all of the 699 SNPs were expected to be imputed with good quality. The squared correlation Rsq metric, which is defined as the observed variance divided by the expected variance, provides a measure of the quality of imputation. The genotypes of 81 SNPs in all subjects and those of the 31 typed SNPs in call-missing subjects were imputed with Rsq≥0.3 (a quality cutoff recommended in the MaCH instruction manual), and those for the control subjects were in Hardy–Weinberg equilibrium (P≥0.05/112=0.00045).
Results
Study design
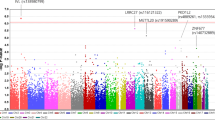
Six SNPs that have recently been associated with POAG susceptibility in a Japanese population29 were typed for all the Korean subjects in this study. Four SNPs (rs540782, rs547984, rs693421, and rs2499601) were located upstream of ZP4 at the 1q43 locus, one SNP (rs7081455) was located far downstream of PLXDC2 at 10p12.31, and one SNP (rs7961953) was located within TMTC2 at 12q21.31. These SNPs were in moderate-to-strong LD with 112 other SNPs (D′≥0.5, r2≥0.1, or D′=1, r2<0.1) in the Japanese population of the International HapMap Project, which could be tagged (r2≥0.7) by 25 SNPs (6 at 1q43, 10 at 10p12.31, and 9 at 12q21.31) in the Korean population of the Korean HapMap Project. All subjects were additionally genotyped for these 25 tag SNPs (Table 2) to identify SNPs that would be in stronger LD in Koreans with causative functional variations than the reported SNPs. Thus, a total of 31 SNPs (ie, the six reported plus 25 tags) in the three loci were genotyped in the initial stage of this study (Table 2).
Replicated strong association of 10p12.31 (downstream of PLXDC2)
Among the 11 typed SNPs at the 10p12.31 locus, a strong association with POAG susceptibility was found for a previously unreported SNP, rs7098387 (Table 2), but the previously reported SNP (rs7081455) showed no association (P=0.74). Only rs7098387 showed a significant association, with a P-value (P=0.0011) lesser than the significance level (α=0.0016) of the Bonferroni correction for multiple testing (Table 2). When imputation was performed with the data for the 11 SNPs, a total of 38 neighboring SNPs were well imputed, though no other SNPs showed a significant association.
The risk effect of the rs7098387 SNP was calculated according to a dominant rather than a recessive or additive genetic model because no case samples were homozygous for the minor allele. The risk was strong (OR=2.0 (1.4–3.0)) for the minor allele (C) carriage in this SNP (P=0.00038). Thus, the individuals carrying the minor allele of rs7098387 had roughly twofold increased odds of having POAG in comparison with respective non-carriers.
To evaluate whether one or more separate association blocks constitute this association locus, conditional association tests were performed for all typed and imputed SNPs, with adjustment for the genotypes of this SNP (ie, rs7098387). All the SNP associations lost significance in the conditional association tests (P≥0.060). Together, these results confirmed the POAG susceptibility association of a locus downstream of PLXDC2 at 10p12.31; however, the risk-associated haplotype was marked by a previously unreported SNP, rs7098387 (allele C), rather than the originally reported rs7081455 (allele T).
Replicated association of 1q43 (harboring ZP4)
Among the 10 typed SNPs of the 1q43 locus, including the four originally reported and six additional tag SNPs, a marginal association was observed with two SNPs, the originally reported rs693421 (P=0.030) and an unreported tag, rs1017982 (P=0.047), according to the co-dominant genetic models (Table 2). When these 10-SNP typing data were used for genotype imputation, 14 SNP genotypes were well imputed, though none showed an association with POAG.
When the risk effect of the reported rs693421 was estimated according to an additive genetic model (less constraint than a dominant or recessive genetic model), the OR value was 1.4 (1.1–1.7) with P=0.0082. The minor allele (T) frequency of the cases was 0.55, which was substantially higher than that of the controls (0.48). Accordingly, a POAG susceptibility association was replicated with rs693421, found within the ZP4-harboring locus of 1q43; however, the association was marginal, most likely due to insufficient statistical power (47% for rs693421).
No association of 12q21.31 (TMTC2 and upstream)
A total of 10 SNPs at the 12q21.31 locus, including an originally reported SNP (rs7961953) and 9 additional tag SNPs, were genotyped (Table 2), and 29 SNPs were imputed in good quality. However, none of these SNPs were associated with POAG susceptibility, either significantly or marginally, in co-dominant genetic models (P>0.05). The tested region covers a 97.3-kb region of the 12q21.31 locus, including a 40-kb 5′-portion of the TMTC2 gene, spanning from rs12822754 (at position 83 040 610) through rs11115376 (at position 83 137 872). The statistical power to detect the originally reported association of rs7961953 was low (49%), most likely explaining the failure to detect POAG association with any of the SNPs at this locus.
Discussion
Three human chromosome loci were recently associated with susceptibility to POAG in a Japanese population,29 but none of the six reported SNP markers of the loci maintained this association in three subsequent studies using South Indian,30 Afro-Caribbean,24 and Chinese31 populations. Rather than merely examining the reported markers for replication in Koreans, the present study aimed to identify non-reported markers of stronger association that could be detected, even with low statistical powers for the reported associations.
POAG susceptibility was discovered to be associated with a non-reported SNP (rs7098387) on chromosome 10p12.31, whereas a reported SNP (rs7081455) was not associated. The newly associated SNP tended to exhibit a stronger effect size (OR=2.0 (1.4–3.0) for rs7098387) than that reported in the Japanese population (OR=1.5 (1.3–1.8) for rs7081455),29 although the 95% confidence interval did overlap. According to the 1000 Genomes Project data, two other SNPs, rs78099346 and rs1412557, were perfectly correlated (r2=1) with the newly associated rs7098387 and are presumably also associated with POAG. These results not only confirm the POAG susceptibility association of the 10p12.31 locus in Koreans but also suggest that future replication studies using other populations should examine rs7098387, rs78099346 or rs1412557 rather than rs7081455.
Among other possible causes, this difference in the association markers between the Korean and Japanese studies can be attributed to differences in the LD structure of the locus between the two populations. The Korean marker rs7098387 was not examined in the original Japanese study; however, according to the 1000 Genomes Project data, the risk-associated allele T of the Japanese marker rs7081455 is in complete LD (D′=1) with the risk-associated allele of rs7098387 (C) in the Japanese population. Accordingly, rs7098387 would presumably exhibit an association in Japanese populations with the same direction of effect. In contrast, rs7081455 was not associated with POAG in Koreans, most likely because it was in low LD (D′=0.40) with rs7098387 in this group.
Therefore, rs7098387 would be valid in both Korean and Japanese populations, whereas rs7081455 would be effective only in Japanese or in case of high statistical power. Because of the low correlation (r2=0.047 in JPT) and despite the complete LD between the two markers, the effect sizes would presumably be different in the Japanese population, as rs7098387 had a larger effect than rs7081455 in the Korean population.
Another possible cause for the different association results of the two studies was the substantial difference in intraocular pressure levels of the glaucoma cases. In this (ie, Korean) study, high-tension cases (88.5% of all cases) were primarily recruited, even though normal-tension glaucoma (2.7%) is much more prevalent than high-tension glaucoma (0.8%) in the general Korean population.37 In addition, the average intraocular pressure was 28.2 mm Hg, which was much higher than the average pressure in the previous Japanese study (15.5 mm Hg).29 Although the effect of the Korean marker rs7098387 on high-tension POAG (OR=2.2 (1.5–3.1), P=0.00012) was not much different from that on total POAG (OR=2.0 (1.4–3.0), P=0.00038) in this study, it remains to be determined whether the rs7098387 and rs7081455 markers are separately associated with high-tension and normal-tension glaucoma, respectively.
The newly identified SNP, rs7098387, is located 54 kb downstream of PLXDC2 (plexin domain containing 2), which encodes a type I transmembrane protein with some homology to the nidogens and plexins that are present in the developing nervous system and may regulate the proliferation of neural progenitors.39, 40 At present, however, no functional significance is implied for this intergenic SNP, though a possibility can be speculated. This SNP is perfectly correlated (r2=1) with rs78099346 and rs1412557, which are, according to the ENCODE project data,41, 42 located in an early-replication domains where chromosomal DNA replication initiates earlier than other chromosomal regions in embryonic stem cells. As the early replication of DNA is closely correlated with high levels of gene transcription and expression,43 it would be worth examining whether these SNPs can affect the expression levels of nearby genes in a long-distance manner.
With regard to replication of the association of the second locus (1q43), one of the four reported SNPs (rs693421) exhibited the lowest P-value (P=0.0082) and the largest effect (OR=1.4 (1.1–1.7)) in additive genetic models (Table 2). A marginal association was detected for rs693421 despite the low statistical power (47%), and the effect size was comparable between the present Korean and reported Japanese (OR=1.4 (1.2–1.6))29 populations. This marginal association most likely supports the replicated association of the 1q43 locus, though the results should be confirmed in larger populations.
The reported association of the third locus (12q21.31) failed to be replicated in this study, possibly because of the low statistical power for the reported rs7961953 (49% for OR=1.4 (1.2–1.6)); therefore, this failure should not be regarded as evidence against any association of the 12q21.31 locus. These results suggested that no neighboring candidate SNPs at 12q21.31 had a much larger effect than the reported SNP (rs7961953) to be detected with a low statistical power.
This study was not designed to provide sufficient statistical power (eg, ≥80%) for the replication of any of the reported SNP markers (power of 47–54%), which were nevertheless genotyped in this study. We additionally examined nearby HapMap SNPs that were in moderate to high LD with the reported SNP markers in the Japanese population. Furthermore, all other SNPs at these loci were imputed according to the Korean HapMap Project data. This strategy covered candidate SNPs of possible indirect association and was effective in identifying novel associations that were even stronger than that reported with respect to the POAG association of the 10p12.31 locus.
In summary, we successfully identified optimal SNP markers that provide the highest statistical power with the largest effect size for replication of the previous Japanese POAG-associated haplotypes at the 10p12.31 and 1q43 loci. Using a Korean population, the POAG association of the 10p12.31 locus was replicated using the previously unreported SNP rs7098387 rather than the previously reported rs7081455, with 1q43 locus association using the reported marker rs693421, in expansive analyses of candidate SNP markers chosen by a combination of LD-based strategy and imputation. Using the replication-optimal markers provided by these findings, association studies of these loci should be facilitated in other populations, particularly non-Asian populations.
Accession codes
References
Kwon YH, Fingert JH, Kuehn MH, Alward WL : Primary open-angle glaucoma. N Engl J Med 2009; 360: 1113–1124.
Anderson DR : Collaborative normal tension glaucoma study. Curr Opin Ophthalmol 2003; 14: 86–90.
Heijl A, Leske MC, Bengtsson B, Hyman L, Hussein M : Reduction of intraocular pressure and glaucoma progression: results from the Early Manifest Glaucoma Trial. Arch Ophthalmol 2002; 120: 1268–1279.
Leske MC, Connell AM, Wu SY, Hyman LG, Schachat AP : Risk factors for open-angle glaucoma. The Barbados Eye Study. Arch Ophthalmol 1995; 113: 918–924.
Tielsch JM, Katz J, Sommer A, Quigley HA, Javitt JC : Family history and risk of primary open angle glaucoma. The Baltimore Eye Survey. Arch Ophthalmol 1994; 112: 69–73.
Doshi V, Ying-Lai M, Azen SP, Varma R : Sociodemographic, family history, and lifestyle risk factors for open-angle glaucoma and ocular hypertension. The Los Angeles Latino Eye Study. Ophthalmology 2008; 115: 639–647 e632.
Weih LM, Nanjan M, McCarty CA, Taylor HR : Prevalence and predictors of open-angle glaucoma: results from the visual impairment project. Ophthalmology 2001; 108: 1966–1972.
Wu J, Hewitt AW, Green CM et al: Disease severity of familial glaucoma compared with sporadic glaucoma. Arch Ophthalmol 2006; 124: 950–954.
Nemesure B, Jiao X, He Q et al: A genome-wide scan for primary open-angle glaucoma (POAG): the Barbados Family Study of Open-Angle Glaucoma. Hum Genet 2003; 112: 600–609.
Wiggs JL, Allingham RR, Hossain A et al: Genome-wide scan for adult onset primary open angle glaucoma. Hum Mol Genet 2000; 9: 1109–1117.
Crooks KR, Allingham RR, Qin X et al: Genome-wide linkage scan for primary open angle glaucoma: influences of ancestry and age at diagnosis. PLoS ONE 2011; 6: e21967.
Porter LF, Urquhart JE, O’Donoghue E et al: Identification of a novel locus for autosomal dominant primary open angle glaucoma on 4q35.1-q35.2. Invest Ophthalmol Vis Sci 2011; 52: 7859–7865.
Stone EM, Fingert JH, Alward WL et al: Identification of a gene that causes primary open angle glaucoma. Science 1997; 275: 668–670.
Rezaie T, Child A, Hitchings R et al: Adult-onset primary open-angle glaucoma caused by mutations in optineurin. Science 2002; 295: 1077–1079.
Monemi S, Spaeth G, DaSilva A et al: Identification of a novel adult-onset primary open-angle glaucoma (POAG) gene on 5q22.1. Hum Mol Genet 2005; 14: 725–733.
Thorleifsson G, Walters GB, Hewitt AW et al: Common variants near CAV1 and CAV2 are associated with primary open-angle glaucoma. Nat Genet 2010; 42: 906–909.
Ramdas WD, van Koolwijk LM, Ikram MK et al: A genome-wide association study of optic disc parameters. PLoS Genet 2010; 6: e1000978.
Macgregor S, Hewitt AW, Hysi PG et al: Genome-wide association identifies ATOH7 as a major gene determining human optic disc size. Hum Mol Genet 2010; 19: 2716–2724.
Burdon KP, Macgregor S, Hewitt AW et al: Genome-wide association study identifies susceptibility loci for open angle glaucoma at TMCO1 and CDKN2B-AS1. Nat Genet 2011; 43: 574–578.
Osman W, Low SK, Takahashi A, Kubo M, Nakamura Y : A genome-wide association study in the Japanese population confirms 9p21 and 14q23 as susceptibility loci for primary open angle glaucoma. Hum Mol Genet 2012; 21: 2836–2842.
Wiggs JL, Kang JH, Yaspan BL et al: Common variants near CAV1 and CAV2 are associated with primary open-angle glaucoma in Caucasians from the USA. Hum Mol Genet 2011; 20: 4707–4713.
Ramdas WD, van Koolwijk LM, Lemij HG et al: Common genetic variants associated with open-angle glaucoma. Hum Mol Genet 2011; 20: 2464–2471.
Burdon KP, Crawford A, Casson RJ et al: Glaucoma risk alleles at CDKN2B-AS1 are associated with lower intraocular pressure, normal-tension glaucoma, and advanced glaucoma. Ophthalmology 2012; 119: 1539–1545.
Cao D, Jiao X, Liu X et al: CDKN2B polymorphism is associated with primary open-angle glaucoma (POAG) in the Afro-Caribbean population of Barbados, West Indies. PLoS ONE 2012; 7: e39278.
Takamoto M, Kaburaki T, Mabuchi A et al: Common variants on chromosome 9p21 are associated with normal tension glaucoma. PLoS ONE 2012; 7: e40107.
Wiggs JL, Yaspan BL, Hauser MA et al: Common variants at 9p21 and 8q22 are associated with increased susceptibility to optic nerve degeneration in glaucoma. PLoS Genet 2012; 8: e1002654.
Mabuchi F, Sakurada Y, Kashiwagi K, Yamagata Z, Iijima H, Tsukahara S : Association between genetic variants associated with vertical cup-to-disc ratio and phenotypic features of primary open-angle glaucoma. Ophthalmology 2012; 119: 1819–1825.
Fan BJ, Wang DY, Pasquale LR, Haines JL, Wiggs JL : Genetic variants associated with optic nerve vertical cup-to-disc ratio are risk factors for primary open angle glaucoma in a US Caucasian population. Invest Ophthalmol Vis Sci 2011; 52: 1788–1792.
Nakano M, Ikeda Y, Taniguchi T et al: Three susceptible loci associated with primary open-angle glaucoma identified by genome-wide association study in a Japanese population. Proc Natl Acad Sci USA 2009; 106: 12838–12842.
Rao KN, Kaur I, Chakrabarti S : Lack of association of three primary open-angle glaucoma-susceptible loci with primary glaucomas in an Indian population. Proc Natl Acad Sci USA 2009; 106: E125–E126, author reply E127.
Chen LJ, Tam PO, Leung DY et al: SNP rs1533428 at 2p16.3 as a marker for late-onset primary open-angle glaucoma. Mol Vis 2012; 18: 1629–1639.
Palmer LJ, Cardon LR : Shaking the tree: mapping complex disease genes with linkage disequilibrium. Lancet 2005; 366: 1223–1234.
Clarke GM, Carter KW, Palmer LJ, Morris AP, Cardon LR : Fine mapping versus replication in whole-genome association studies. Am J Hum Genet 2007; 81: 995–1005.
Konig IR : Validation in genetic association studies. Brief Bioinform 2011; 12: 253–258.
Kraft P, Zeggini E, Ioannidis JP : Replication in genome-wide association studies. Stat Sci 2009; 24: 561–573.
Kim K, Yun YJ, Kim S, Kim JS, Kim CS, Kang C : Analysis of an extended chromosome locus 2p14-21 for replication of the 2p16.3 association with glaucoma susceptibility. Mol Vis 2011; 17: 1136–1143.
Kim CS, Seong GJ, Lee NH, Song KC : Prevalence of primary open-angle glaucoma in central South Korea the Namil study. Ophthalmology 2011; 118: 1024–1030.
Ju H, Cho H, Kim YS et al: Functional polymorphism 57Val>Ile of aurora kinase A associated with increased risk of gastric cancer progression. Cancer Lett 2006; 242: 273–279.
Miller SF, Summerhurst K, Runker AE et al: Expression of Plxdc2/TEM7R in the developing nervous system of the mouse. Gene Expr Patterns 2007; 7: 635–644.
Miller-Delaney SF, Lieberam I, Murphy P, Mitchell KJ : Plxdc2 is a mitogen for neural progenitors. PLoS ONE 2011; 6: e14565.
ENCODE Project Consortium: The ENCODE (ENCyclopedia Of DNA Elements) Project. Science 2004; 306: 636–640.
Birney E, Stamatoyannopoulos JA, Dutta A et al: Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 2007; 447: 799–816.
Goren A, Cedar H : Replicating by the clock. Nat Rev Mol Cell Biol 2003; 4: 25–32.
Acknowledgements
The authors thank Dr Yong-jun Yun for assistance in the blood sample collection and Ms Yun Yeong Moon for administrative and technical assistance. This work was partially supported by grants from National Research Foundation of Korea (2011-0017999 and 2011-0020334).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Kim, K., Heo, D., Kim, S. et al. Expansive marker analysis replicating the association of glaucoma susceptibility with human chromosome loci 1q43 and 10p12.31. Eur J Hum Genet 22, 409–413 (2014). https://doi.org/10.1038/ejhg.2013.149
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2013.149
Keywords
This article is cited by
-
Conserved sequence motifs in human TMTC1, TMTC2, TMTC3, and TMTC4, new O-mannosyltransferases from the GT-C/PMT clan, are rationalized as ligand binding sites
Biology Direct (2021)
-
Polymorphisms rs693421 and rs2499601 at locus 1q43 and their haplotypes are not associated with primary open-angle glaucoma: a case–control study
BMC Research Notes (2019)
-
Plexin domain containing 2 (PLXDC2) gene polymorphism rs7081455 may not influence POAG risk in a Saudi cohort
BMC Research Notes (2018)
-
Polymorphism rs547984 on human chromosome 1q43 is not associated with primary open angle glaucoma in a Saudi cohort
Journal of Negative Results in BioMedicine (2017)