Abstract
We report a novel pathogenic mutation of the mitochondrial transfer RNA (tRNA) gene for tryptophan in a patient with isolated myopathy and persistently elevated creatine kinase. Muscle studies revealed ragged red fibres and decreased activity of respiratory chain complex I and cytochrome c oxidase (COX). Sequencing of the 22 mitochondrial tRNA genes revealed a mutation m.5522G>A, which alters a conserved base pairing in the D-stem of the tRNA for tryptophan. The mutation was heteroplasmic with a mutational load between 88 and 99% in COX-negative fibres. This case contributes to the genetic heterogeneity of mitochondrial diseases caused by mutations in mitochondrial tRNA genes.
Similar content being viewed by others
Introduction
Human mitochondrial DNA (mtDNA) is a circular double-stranded molecule of 16 569 bp, which contains 37 genes. This genome encodes 2 ribosomal RNAs, 22 transfer RNAs (tRNA) and 13 polypeptides that are subunits of respiratory chain complexes. MtDNA is continuously turned over independently of the nuclear genome, but for mtDNA replication and transcription, nuclear-encoded factors are indispensable.1 The mitochondrial genome has a very high mutation rate that is attributable to the lack of protective histone proteins, inefficient repair systems and the vicinity of respiratory chain complexes and reactive oxygen species that they produce.2 More than 92% of the mtDNA sequence encodes genes and the remaining small non-coding areas contain regulatory sequences for replication and transcription, hence almost any nucleotide substitution could have phenotypic effect.3 The majority of described mutations are neutral polymorphisms, which contribute to diversity of human population.4 Pathogenic mutations in the mtDNA can be classified as large-scale rearrangements, point mutations in protein-coding genes or point mutations in tRNA genes.5 Owing to the central role of mitochondria in the cell energy production, phenotypic expression is most likely to occur in tissues with high-energy requirements such as central nervous system, skeletal muscle and heart.6 To date, 549 point mutations of the mtDNA with reports of disease association have been listed in the mitomap database.7 More than half of disease-related point mutations are located within mitochondrial tRNA genes that comprise only ≈9% of the mitochondrial genome, therefore we can assume that these sequences are hot spots for mitochondrial pathogenesis.8 Mutations in tRNA genes usually cause multisystem disorders, including mitochondrial encephalomyopathy, lactic acidosis and stroke-like episodes – the most common maternally inherited mitochondrial disorder.9 The same pathogenic mtDNA mutation may give rise to different phenotypes and mutation carriers may be asymptomatic or express a wide spectrum of symptoms varying in clinical severity.10 The clinical expression of mtDNA mutations depends on the percentage of heteroplasmy, tissue energy demand and the type of mutation.
The biochemical phenotype of tRNA mutations is a lower cellular oxygen consumption, lower enzyme activities of respiratory chain complexes comprises mtDNA-encoded subunits (often decreased activity of complex I and complex IV) and lower ATP synthesis.11
In this report, we describe a novel mitochondrial tRNA mutation in the mitochondrial tRNA gene for tryptophan (MT-TW gene) in a patient with mild muscle symptoms and persistently elevated creatine kinase (CK).
Patients and methods
Case report
The patient is the only child of unrelated parents. Family history was negative for both muscle disease and other health problems, which may indicate presence of the described mutation in relatives. The patient’s psychomotor development was normal. During childhood, he had frequent tonsillitis, allergic rhinitis and occasional headaches, but otherwise was healthy. At the age of 17 years, he was admitted to the hospital following 2 weeks of oppressive feeling in the chest, dry cough and low-grade fever. Physical examination and lung function tests showed normal findings. Standard laboratory workup revealed high CK values (724–1850 U/l) and elevated urine myoglobine (86.4 ng/l; NV <30). The results of other biochemical investigations including lactate, ammonia, carnitine, viral antibodies and autoimmunity tests were all within the reference range. ECG and heart ultrasound were normal. Multiplex PCR of dystrophin gene showed no deletion. During hospitalization, the patient was treated with carnitine (3 × 1 g) for 10 days. He was discharged in good general condition without the symptoms, but with elevated CK of 1031 U/l. Subsequently, he occasionally complained of the muscle pain in the chest and extremities, usually in the morning.
Two years later, he was admitted to our outpatient clinic because of permanently elevated CK (up to 2500 U/l, 98.1% – MM isoform) and electromyography (EMG) signs of mild myopathy in brachial muscles. Muscle bulk and strength were not reduced, and the physical examination showed normal results. Thyroid function tests were normal. Metabolic work-up showed normal urinary organic acids, serum total and free carnitine, acylcarnitine profile, and alpha-glucosidase activity in lymphocytes. Amino-acid analysis showed mildly elevated plasma isoleucine and a generalized increase in urinary amino-acid excretion, which was not associated with any other sign of tubulopathy. Plasma creatinine and electrolytes were normal. Forearm ischaemic test pointed neither to muscle glycogenoses nor to myoadenylate deaminase deficiency.
A deltoid muscle biopsy was performed and both the histopathological and ultrastructural analysis showed the signs of mitochondrial myopathy, which was confirmed both by finding decreased activity of complexes I, III, IV and V of the respiratory chain and the mutation m.5522G>A of the MT-TW gene (see Results section). After establishing the diagnosis of mitochondrial myopathy, vitamins B1, B2 and C were recommended. Owing to limited compliance, the follow-up was very irregular and our plans to monitor particularly muscle and kidney have not been feasible. Despite our recommendations to avoid hard exercises, the patient practiced competitive sports.
As mtDNA mutations are transmitted by maternal inheritance and sporadic cases are rarely reported,12 after confirming mutation in mitochondrial tRNA gene in the patient, his mother of 45 years was also checked. She had no health complaints, the neurological examination was normal, yet her EMG revealed mild generalized myopathy and she had elevated plasma CK of 266 U/l (normal for age <135) and myoglobine 69 μg/l (reference range 19–51) suggesting that her muscles are affected, likely due to the mtDNA mutation detected in her son. So far, she did not allow muscle biopsy.
Muscle histopathology
Muscle histochemistry included the stainings for nicotinamide adenine dinucleotide (NADH), succinate dehydrogenase (SDH) and modified Gomori’s trichrome. Additional double SDH plus cytochrome c oxidase (COX) staining was performed on frozen muscle sections.13 For immunohistochemistry, antibodies against dystrophin, spectrin, emerin, alpha-sarcoglycan and merosin were used. Ultrastructural analysis was performed on Philips Morgagni 268D transmission electron microscope (Hillsboro, OR, USA).
Biochemical analysis in muscle tissue
Fresh muscle samples were taken in the operating room and a part of the specimen was immediately snap frozen in liquid nitrogen. The sample was stored at −70 °C until shipment. Biochemical analysis of the electron transport chain complexes I, II, III, IV, oligomycin-sensitive ATPase, pyruvate dehydrogenase and citrate synthase activity were performed on 600 g supernatant of frozen muscle homogenate.14
DNA studies
Genomic DNA was extracted from the muscle specimen and urine sediment by proteinase K digestion at 60 °C for 2 h and from blood by the use of an extraction kit (NucleoSpin Blood, Machery-Nagel, Düren, Germany). Amplification of the mtDNA was performed as reported.15 Sequence analysis was done on a CEQ-8000 system (Beckman Coulter, Vienna, Austria) according to the manufacturer’s instructions. Single fibres were dissected under the microscope from COX/succinate dehydrogenase double-stained histochemical sections (Figure 1a). DNA was extracted and used for PCR amplification.13 A mutation selective PCR reaction was performed with the forward primer 5′-CACCATCATAGCCACCATCA-3′ and the reverse primer 5′-TGAAGGCTCTTGGTCTGTATTTGA-3′, which carries a mismatch at the underlined position to generate a restriction site for the restriction enzyme MboI in case of the mutation m.5522A>G. A fluorescently (Cy-5) labelled forward primer was added before the last PCR cycle. Ten microlitres of the labelled PCR product were digested by the addition of five units MboI (Thermo Scientific, St Leon-Rot, Germany) diluted in 10 μl reaction buffer and incubation for 16 h at 37 °C. Five microlitres were loaded on a capillary electrophoresis apparatus (CEQ-8000).
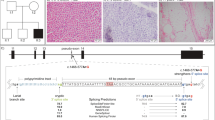
(a) COX plus SDH double staining of frozen muscle section showing numerous COX-negative and SDH-positive fibres in blue. The proportion of COX-positive fibres was 41%, 27% were COX-negative and 32% of the fibres were stained in an intermediate intensity. (b) Electron microscopic figure showing two skeletal muscle fibres, the one on the left with subsarcolemmal accumulation of enlarged and abnormally shaped mitochondria (arrow) (magnification × 8900).
Results
Muscle histopathology
Light microscopy showed normal cytoarchitecture of skeletal muscle with mild variation in size of muscle fibres. NADH and SDH staining demonstrated few fibres showing subsarcolemmal accumulation of oxidative enzyme activity. Modified Gomori trichrom staining revealed single ragged red fibres. Additional COX plus SDH double staining of frozen muscle section showed numerous COX-negative and SDH-positive fibres (Figure 1a). Immunohistochemistry showed normal expression of dystrophin, spectrin, emerin, alpha-sarcoglycan and merosin. Electron microscopy revealed individual muscle fibres with subsarcolemmal aggregates of abnormally shaped mitochondria (Figure 1b).
Biochemical analysis in muscle tissue
Investigation of respiratory chain complex activities showed combined deficiency of complex I and to a milder extent of complexes III, IV and V (Table 1), which confirmed the diagnosis of mitochondrial disease. The activity of the fully nuclear-encoded enzymes citrate synthase and complex II were above the normal range when compared with the protein content, which indicates a compensatory upregulation and is in line with the results of the histological investigations.
DNA studies
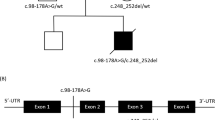
Muscle morphology and result of enzyme assays indicated a defect either within mtDNA or in mitochondrial replication, transcription and translation. Therefore, sequence analysis of mitochondrial tRNA genes was performed and revealed a mutation m.5522G>A, which is located within the MT-TW gene of the mitochondrial tRNA for tryptophan (Figure 2). The mutation was heteroplasmic, the mutation load was 76% in DNA from muscle homogenate, 11% in DNA from urine and 5% in DNA from the blood of the patient. The mutation was not detectable in the DNA extracted neither from the blood nor from the urine of the mother. Analysis of DNA extracted from single muscle fibres, which were either COX-positive or -negative, revealed a clearly higher mutation load in the COX-negative fibres (Figure 3a).
Mutation analysis revealed a mutation m.5522G>A (a). Restriction fragment length analysis (b) revealed a mutation load of 76% in the patient muscle and 5% in the patient blood. The mutation was not detectable in the blood of the patient’s mother. Cloverleaf structure of the human mitochondrial tRNATrp gene (c). Known pathogenic mutations are indicated in red. Sequence variations published in controls either in the mitomap database7 (green), the Human Mitochondrial Genome Database (http://www.mtdb.igp.uu.se/) (blue) or in both (purple) are indicated in colours. The novel mutation found in our patient is bolded. r.f.u., relative fluorescence units.
Quantification of the mutation m.5522G>A in single muscle fibres dissected from COX/SDH double-stained histochemical muscle preparations (a). COX-positive (n=14): mean 17%, SD=18%, min 2%, max 57%; COX-negative (n=17): mean 96%, SD=3%, min 88%, max 99%; Student’s unpaired t-test: P<0.0001. The affected position in the D-arm of the tRNATrp is conserved within Metazoans (b). The arrows indicate the affected base pairs in the D-stem of the tRNATrp.
Discussion
We identified a novel pathogenic mtDNA point mutation in the region specifying for the D-stem of mitochondrial tRNATrp. Our patient exhibited discrete EMG signs of myopathy, and occasional muscle pain without fatigue or symptomatic weakness. The only persistent sign of muscle disease was moderately high CK values. The mechanism of CK elevation in this patient is probably, at least partly, due to muscle damage as it can be concluded by parallel increase in plasma myoglobin. Another contributing factor, when ATP synthesis is impaired, could be activation of AMP-activated protein kinase, an energy sensing enzyme, which could lead to expulsion of CK from the cytosol.16 Isolated myopathy with proximal limb weakness with fatigue and slowly deterioration was reported as a frequent manifestation of mtDNA disorders.17 A number of mitochondrial tRNA point mutations have been associated with isolated mitochondrial myopathy.18 The subsarcolemmal accumulation of mitochondria and presence of ragged red fibres, as found in our patient, are indicative for alterations of functional proteins and are associated with mitochondrial tRNA mutations.19
Biochemical analysis revealed a combined deficiency in complexes I and IV. As this constellation was typical for a defect either within mtDNA or in mitochondrial replication, transcription and translation, sequencing of all tRNA genes was performed and identified a new point mutation in the tRNATrp gene. The mutational load varied between 5% in DNA from blood and 76% in DNA from muscle homogenate. Heteroplasmy with a clear predominance of the mutated genome in COX-negative muscle fibres indicated that this mutation is pathogenic, according also to the scoring system for assessing pathogenicity proposed by Yarham et al.20 Cell culture studies performed by Boluet et al21 on myoblast culture from MERRF patients revealed a threshold effect for mitochondrial translation, which remains unaffected by tRNA point mutation until <85% of the mtDNA molecules are mutated.
To our knowledge, the mutation 5522G>A within the MT-TW gene of the mitochondrial tRNA for tryptophan has not been reported previously and was not observed in normal controls. The mutation is located in a conserved region; in most animal mitochondrial tRNATrp there is always a guanosine with a perfect base-pairing found at this position of the D-stem (Figure 3).22 It affects a double bond in the D-stem of the tRNA where two other pathogenic mutations have been described in neighbouring nucleotides (Figure 2c). Silvestri et al.23 reported of G>A transition at position 5521 of mitochondrial tRNATrp gene in a male patient with late onset myopathy. The biochemical phenotype expressed by that patient was a selective decrease in COX activity and authors suggested two pathogenic mechanisms that could synergistically act to decrease the rate of synthesis of COX subunits. The first is the site of mutation, located in the D-stem, which can affect tRNA–ribosomal interaction with a consequent decrease in mitochondrial protein synthesis, and the second is higher relative content of tryptophan in two catalytic subunits of COX (COI and COIII) compared with other mtDNA-encoded respiratory chain subunits (Supplementary Table 1). A mutation in another neighbouring nucleotide, at position 5523 of mtDNA was reported in a 13-year-old patient with Leigh syndrome. The T5523G mutation was detected in heteroplasmic form and authors showed that this alteration of tRNATrp gene leads to a disruption of the secondary structure of tRNATrp.24 Relatively mild phenotype associated with the mutation found in our patient in comparison with the Leigh encephalopathy in a patient with the neighbouring 5523T>G mutation could be due to different effects on secondary structure of tRNATrp, but also due to different mutation load in the brain tissue. Other pathogenic mt-tRNATrp mutations (Figure 2c, Table 2) are associated with multisystem mitochondrial disorders, usually with predominant neurological symptoms with myopathy and encephalopathy as the most frequent clinical manifestation.24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35 A possible pathogenic effect of disease-related tRNA mutations is their impact on tRNA conformation. Structural characteristics of tRNA are a cloverleaf-like secondary structure and ‘L’-shaped three-dimensional structure. A single-nucleotide substitution can destabilize the secondary and tertiary structure of a tRNA and result in destabilization and tRNA impaired function.11 Most of the pathological tRNA mutations occur in the stem structure.8
This report of a novel mt-tRNATrp mutation further highlights the role of mitochondrial tRNA alterations as a possible cause of mitochondrial myopathy.
References
Taanman JW : The human mitochondrial genome: structure, transcription, translation and replication. Biochym Biophys Acta 1999; 1410: 103–123.
Tuppen H, Blakely EL, Turnbull DM, Taylor RW : Mitochondrial DNA mutations and human disease. Biochimica Biophys Acta 2010; 1797: 113–128.
Montoya J, Lopez-Gallardo E, Diez-Sanchez C, Lopez-Perez MJ, Ruiz-Pesini E : 20 years of human mtDNA pathologic point mutations: carefully reading the pathogenicity criteria. Biochimica Biophys Acta 2009; 1787: 476–483.
Ingman M, Kaessermann H, Pääbo S, Gyllenstein U : Mitochondrial genome variation and the origin of modern humans. Nature 2000; 408: 708–713.
Crimi M, Rigolio F : The mitochondrial genome, a growing interest inside as organelle. Int J Nanomedicine 2008; 3: 51–57.
Chinnery PF, Howell N, Andrews RM, Turnbull DM : Clinical mitochondrial genetics. J Med Genet 1999; 36: 425–436.
MITOMAP: a human mitochondrial genome database http://www.mitomap.org, 2012.
Scaglia F, Wong LJ : Human mitochondrial transfer RNAs: role of pathogenic mutation in disease. Muscle Nerve 2008; 37: 150–171.
Servidei S : Mitochondrial encephalomyopathies: gene mutation. Neuromuscul Disord 2003; 13: 848–853.
Kaufmann P, Engelstad K, Wei Y et al: Protean phenotypic features of the A3243G mitochondrial DNA mutation. Arch Neurol 2009; 66: 85–91.
Florentz C, Sohm B, Tryoen-Toth P, Pütz J, Sissler M : Human mitochondrial tRNAs in health and disease. Cell Mol Life Sci 2003; 60: 1356–1375.
Thorburn DR, Dahl HH : Mitochondrial disorders: genetics, counseling, prenatal diagnosis and reproductive options. Am J Med Genet 2001; 106: 102–114.
Oldfors A, Moslemi AR, Fyhr IM, Holme E, Larsson NG, Lindberg C : Mitochondrial DNA deletions in muscle fibers in inclusion body myositis. J. Neuropathol Exp Neurol 1995; 54: 581–587.
Mayr JA, Paul J, Pecina P et al: Reduced respiratory control with ADP and changed pattern of respiratory chain enzymes as a result of selective deficiency of the mitochondrial ATP synthase. Pediatr Res 2004; 55: 988–994.
Meierhofer D, Mayr JA, Ebner S, Sperl W, Kofler B : Rapid screening of the entire mitochondrial DNA for low-level heteroplasmic mutations. Mitochondrion 2005; 5: 282–296.
Baird MF, Graham SM, Baker JS, Bickerstaff GF : Creatine-kinase- and exercise-related muscle damage implications for muscle performance and recovery. J Nutr Metab 2012; 2012: 960363.
Leonard JV, Schapira AVH : Mitochondrial respiratory chain disorders I: mitochondrial DNA defects. Lancet 2000; 355: 299–304.
Kleinle S, Schneider V, Moosmann P, Brandner S, Krähenbühl S, Liechti-Gallati S : A novel mitochondrial tRNAPhe mutation inhibiting anticodon stem formation associated with a muscle disease. Biochem Biophys Res Commun 1998; 247: 112–115.
Filosto M, Tomelleri G, Tonin P et al: Neuropathology of mitochondrial diseases. Biosci Rep 2007; 27: 23–30.
Yarham JW, Al-Dosary M, Blakely EL et al: A comparative analysis approach to determining the pathogenicity of mitochondrial tRNA mutations. Hum Mutat 2011; 32: 1319–1325.
Boluet L, Karpati G, Shoubridge EA : Distribution and threshold expression of the tRNA(Lys) mutation in skeletal muscle of patients with myoclonic epilepsy and ragged-red fibers (MERRF). Am J Hum Genet 1992; 51: 1187–1200.
Mamit-tRNA: compilation of mammalian mitochondrial tRNA genes http://mamit-tRNA.u-strasbg.fr/.
Silvestri G, Rana M, DiMuzio A, Uncini A, Tonali P, Servidei S : A late onset mitochondrial myopathy is associated with a novel mitochondrial DNA (mtDNA) point mutation in the tRNATrp gene. Neuromuscul Disord 1998; 8: 291–295.
Mkaouar-Rebai E, Chamkha I, Kammoun F et al: Two new mutations in the MT-TW gene leading to the disruption of the secondary structure of the tRNATrp in patients with Leigh syndrome. Mol Genet Metab 2009; 97: 179–184.
Maniura-Weber K, Taylor RW, Johnson MA et al: A novel point mutation in the mitochondrial tRNA(Trp) gene produces a neurogastrointestinal syndrome. Eur J Hum Genet 2004; 12: 509–512.
Santorelli FM, Tanji K, Sano M et al: Maternally inherited encephalopathy associated with a single-base insertion in the mitochondrial tRNATrp gene. Ann Neurol 1997; 42: 256–260.
Malfatti E, Cardaioli E, Battisti C et al: A novel point mutation in the mitochondrial tRNA(Trp) gene produces late-onset encephalomyopathy, plus additional features. J Neurol Sci 2010; 297: 105–108.
Silvestri G, Mongini T, Odoardi F et al: A new mtDNA mutation associated with a progressive encephalopathy and cytochrome c oxidase deficiency. Neurology 2000; 54: 1693–1696.
Anitori R, Manning K, Quan F et al: Contrasting phenotypes in three patients with novel mutations in mitochondrial tRNA genes. Mol Genet Metab 2005; 84: 176–188.
Sacconi S, Salviati L, Nishigaki Y et al: A functionally dominant mitochondrial DNA mutation. Hum Mol Genet 2008; 17: 1814–1820.
Nelson I, Hanna MG, Alsanjari N, Scaravilli F, Morgan-Hughes JA, Harding AE : A new mitochondrial DNA mutation associated with progressive dementia and chorea: a clinical, pathological, and molecular genetic study. Ann Neurol 1995; 37: 400–403.
Smits P, Mattijssen S, Morava E et al: Functional consequences of mitochondrial tRNA Trp and tRNA Arg mutations causing combined OXPHOS defects. Eur J Hum Genet 2010; 18: 324–329.
Sanaker PS, Nakkestad HL, Downham E, Bindoff LA : A novel mutation in the mitochondrial tRNA for tryptophan causing a late-onset mitochondrial encephalomyopathy. Acta Neurol Scand 2010; 121: 109–113.
Valente L, Piga D, Lamantea E et al: Identification of novel mutations in five patients with mitochondrial encephalomyopathy. Biochim Biophys Acta 2009; 1787: 491–501.
Tulinius M, Moslemi AR, Darin N et al: Leigh syndrome with cytochrome-c oxidase deficiency and a single T insertion nt 5537 in the mitochondrial tRNATrp gene. Neuropediatrics 2003; 34: 87–91.
Acknowledgements
This work was supported by Ministry of Science, Education and Sport, Republic of Croatia (grant no.: 108-1081870-1885) and the E-rare 2 (European Research Projects on Rare Diseases) ‘Mitochondriopathy—Network for diagnostics and therapy (GENOMIT)’ financed by the FWF project number I 920-B13.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Barić, I., Fumić, K., Petković Ramadža, D. et al. Mitochondrial myopathy associated with a novel 5522G>A mutation in the mitochondrial tRNATrp gene. Eur J Hum Genet 21, 871–875 (2013). https://doi.org/10.1038/ejhg.2012.272
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2012.272
Keywords
This article is cited by
-
Complex multisystem phenotype associated with the mitochondrial DNA m.5522G>A mutation
Neurological Sciences (2019)