Abstract
Rett syndrome is a clinically defined neurodevelopmental disorder almost exclusively affecting females. Usually sporadic, Rett syndrome is caused by mutations in the X-linked MECP2 gene in ∼90–95% of classic cases and 40–60% of individuals with atypical Rett syndrome. Mutations in the CDKL5 gene have been associated with the early-onset seizure variant of Rett syndrome and mutations in FOXG1 have been associated with the congenital Rett syndrome variant. We report the clinical features and array CGH findings of three atypical Rett syndrome patients who had severe intellectual impairment, early-onset developmental delay, postnatal microcephaly and hypotonia. In addition, the females had a seizure disorder, agenesis of the corpus callosum and subtle dysmorphism. All three were found to have an interstitial deletion of 14q12. The deleted region in common included the PRKD1 gene but not the FOXG1 gene. Gene expression analysis suggested a decrease in FOXG1 levels in two of the patients. Screening of 32 atypical Rett syndrome patients did not identify any pathogenic mutations in the PRKD1 gene, although a previously reported frameshift mutation affecting FOXG1 (c.256dupC, p.Gln86ProfsX35) was identified in a patient with the congenital Rett syndrome variant. There is phenotypic overlap between congenital Rett syndrome variants with FOXG1 mutations and the clinical presentation of our three patients with this 14q12 microdeletion, not encompassing the FOXG1 gene. We propose that the primary defect in these patients is misregulation of the FOXG1 gene rather than a primary abnormality of PRKD1.
Similar content being viewed by others
Introduction
Rett syndrome is a severe neurodevelopmental disorder caused by mutations in the X-linked gene encoding the methyl-CpG-binding protein, MECP2. In the classic form of Rett syndrome (MIM 312750), females have psychomotor regression, profound intellectual impairment, seizures and stereotypic hand movements.1 Usually sporadic, Rett syndrome is caused by mutations in the X-linked MECP2 gene in ∼90–95% of classic cases and 40–60% of individuals with atypical Rett syndrome.2 A number of Rett syndrome variants have been described including the congenital, the early-onset seizure and the preserved speech variants. Mutations in the CDKL5 gene have been associated with the early-onset seizure variant of Rett syndrome, although the predominant phenotype is an epileptic encephalopathy.3, 4, 5 The congenital variant is characterised by infantile hypotonia and developmental delay evident earlier than in classic Rett syndrome but causative mutations have previously rarely been identified in these patients.6
Recent reports of microdeletions in the 14q12 region found in Rett syndrome patients have found a third gene, forkhead box protein G1 (FOXG1), to be highly associated with the congenital variant.7, 8 The FoxG1 protein is a winged-helix transcription factor and its expression is highest in fetal and infant brain. It is involved in telencephalic development,9 promotes neurogenesis and antagonises neuronal differentiation.10, 11 At least eight instances of 14q12 microdeletions have been reported.7, 8, 12, 13, 14 Mutation analyses have identified de novo loss-of-function point mutations in FOXG1, particularly in patients with the congenital form of Rett syndrome.15, 16, 17 Duplications of FOXG1 are also found in patients with epilepsy and intellectual impairment,18, 19, 20, 21 highlighting the importance of gene dosage at this locus, although more recently the pathogenicity of FOXG1 duplications has been questioned.22
We report the clinical features and array CGH findings of three atypical Rett syndrome patients. The three patients, two female and one male, all had intellectual impairment, early-onset developmental delay without regression, microcephaly and hypotonia. In addition, the females had a seizure disorder and agenesis of the corpus callosum. All three were found to have an interstitial deletion of 14q12. The commonly deleted region included the PRKD1 gene but not the FOXG1 gene.
Materials and methods
Case report 1
We first reviewed this girl at age 7 years and 8 months. She was referred for an opinion regarding the possible underlying diagnosis of Rett syndrome as a cause of developmental delay, microcephaly, hypotonia and seizure disorder. The girl was the second child to non-consanguineous parents. She was born by normal vaginal delivery at 42 weeks of gestation, after a normal pregnancy. Her birth weight was 3.46 kg (25–50th percentile), length was 53 cm (50–75th percentile) and head circumference was 33.5 cm (8th percentile). Developmental milestones were delayed. She rolled at 4 months, but was not able to sit without support until ∼1 year of age and never learnt to walk. She was never able to hold a bottle, feed herself and did not develop a pincer grip. Her parents reported that she constantly had her hands in her mouth. Her hearing was normal. At 1 year of age she developed infantile spasms. A gastrostomy was placed for poor weight gain and feeding difficulties. She suffered with constipation, had bruxism and a disturbed sleep pattern.
Repeated examinations over time demonstrated subtle dysmorphic features, including an oval-shaped face, depressed nasal bridge, anteverted nares, a full and everted lower lip and limited facial expression with a tendency to hold the mouth open. At 7 years and 8 months of age, her weight was 14 kg (<3rd percentile) and head circumference was 43.5 cm (<<1st percentile). She demonstrated mid-line hand grasping and hand wringing movements, tongue thrusting and hand mouthing. She had a left esotropia, small, cold hands and feet with relatively long fingers and toes, left second toe clinodactyly and shortening of the 4th and 5th metatarsals. There was one café-au-lait lesion over the right buttock and she had a relatively hirsute back. She had truncal hypotonia and was unable to sit without support. She had mild limb hypertonia and brisk deep tendon reflexes, which over time developed into spastic quadriparesis associated with fixed flexion deformities. Her seizures continued, becoming tonic–clonic, and were associated with choreoathetoid movements. She subsequently developed a thoracolumbar scoliosis and a disturbed breathing pattern with episodes of hyperventilation. She was noted to have reduced bone mineral density and was treated with pamidronate infusions.
A cerebral MRI scan revealed agenesis of the corpus callosum and relatively small brain size. Karyotype, methylation studies for Angelman syndrome, subtelomere FISH studies, MECP2 mutation testing, including MLPA studies and CDKL5 analysis, were normal. In addition, urinary amino and organic acid analyses, plasma lactate, pyruvate, ammonia, transferrin isoforms, very long chain fatty acids and TORCH serology were normal. Array CGH studies detected a 1.17-Mb deletion of 14q12. Parental studies were normal. See ‘Chromosomal microarray analysis methods’ for procedural details.
The patient died at age 16 years from an acute respiratory illness complicating a slow deterioration in her general health.
Case report 2
This 11-year-old girl was referred for assessment of profound global developmental delay, microcephaly, truncal hypotonia, seizures and hand stereotypies. She was born after a normal pregnancy and vaginal delivery at 38 weeks of gestation. Her birth weight was 2.6 kg (9th percentile), length was 46 cm (6th percentile) and head circumference was 31 cm (1st percentile). She was hypotonic from birth, resulting in some initial feeding difficulties. At 3 months, she was noted to have a convergent strabismus and tongue thrusting movements. All developmental milestones were significantly delayed – she smiled at 2 months, rolled at 14 months and sat unsupported in her second year. She never learnt to walk. She reaches for an object but her hand function was restricted to swiping at objects and she has not developed a pincer grip. She had limited babbled only. A formal assessment at age 13 months indicated profound developmental delay in all areas (Griffith Mental Developmental Scale). Her general health and appetite were good but she was not able to feed herself and suffered with constipation. She had a disturbed sleep pattern, episodes of deep breathing, tongue thrusting and hand mouthing movements. At 11 years of age she developed a seizure disorder. The EEG demonstrated occasional centro-parietal epileptiform activity on a slow background and hence she was commenced on carbamazepine.
On examination at 11 years and 9 months, her weight was 39 kg (40th percentile), length was 142 cm (20th percentile) and head circumference was 50.5 cm (<1st percentile). She had truncal hypotonia, limb hypertonia and brisk deep tendon reflexes. There was no scoliosis. She demonstrated midline hand grasping, hand wringing and hand-to-mouth movements. She had an alternating esotropia corrected with spectacles for hypermetropia. She had relatively long tapered fingers and toes (75th centile). Her facial features were subtly different to her parents with microcephaly, long palpebral fissures, slightly upturned palpebral fissures, short upturned nose, cupped ears with simple thickened helices and mouth held open. She had a laughing ‘happy’ disposition and startled easily to noise, movement or light stimulation.
A cerebral MRI scan revealed slight prominence of the body of the lateral ventricles, especially posteriorly and agenesis of the corpus callosum. MECP2 gene sequencing and deletion screening (MLPA studies) were normal. Normal results were obtained for other investigations including a karyotype, transferrin isoforms, methylation studies for Angelman syndrome (SNRPN1 methylation) and urinary glycosaminoglycans, amino and organic acids. Chromosome microarray detected a 1.15-Mb deletion in chromosome 14q12. Parental studies indicated this was de novo.
Case report 3
We first saw this boy at age 6 months, when he was referred for an opinion regarding developmental delay and microcephaly. He was the second child to non-consanguineous parents. A left-sided pelvi-ureteric junction obstruction was detected antenatally. The mother carried a known balanced translocation (46, XX, t(12;21)(p11.2;p11.2) and the boy’s karyotype, performed by amniocentesis, was reported as normal. He was born at term by caesarean section. His birth weight was 4.4 kg (97th percentile), length was 52.5 cm (50–75th percentile) and head circumference was 34 cm (14th percentile). Postnatally, he required surgery for the pelvi-ureteric junction obstruction. By 6 months of age, he had developed microcephaly with a head circumference of 39.7 cm (<3rd percentile) and positional plagiocephaly. He was nondysmorphic and his other growth parameters were normal; weight was 7.7 kg (50th percentile) and height was 68 cm (50th percentile). His fine and gross motor developmental milestones were delayed, although he had good social development. He had poor use of his hands. He had increased muscle tone in all limbs with brisk deep tendon reflexes in his lower limbs. Hearing and vision were normal. Following several apnoeic episodes at 10 months of age, he underwent a formal sleep study (polysomnography), which showed mixed central and obstructive sleep disordered breathing. There was progressive deceleration of head growth; at 1 year of age, his head circumference was 41.5 cm (>2 cm below the 2nd percentile). By 4 years of age, he had severe developmental delay. He had learnt to crawl and stand with support. His hand function was restricted to picking up objects and finger feeding. He had minimal speech, limited to a few single words and had learnt some sign language. He had difficulty feeding with tongue thrusting movements and gagging. He also had bruxism and cold feet. He suffered with constipation, had a disturbed sleep pattern with night laughter but had never had a seizure. He had midline hand grasping, hand tapping and hand-to-mouth movements. He had a happy disposition and startled easily.
Postnatally, a karyotype and TORCH screen were normal. Subsequently, at 2 years of age, array CGH detected a 1.80-Mb deletion of 14q12. Parental studies were normal.
The clinical features of all three patients are summarised in Table 1.
Sample collection
Patient samples were received for clinical diagnostic testing and parental samples were subsequently obtained. Informed consent was obtained from the parents of the patients.
Chromosomal microarray analysis methods
For all three patients, CGH microarray analysis was performed at the Cytogenetics Department at The Children’s Hospital at Westmead, using Agilent Sureprint G3Hmn 400K arrays (Agilent Technologies, Santa Clara, CA, USA), with data analysed at an effective median resolution of 0.06 Mb. Arrays were handled and loaded as per the manufacturer’s specifications [http://www.genomics.agilent.com/]. Array scans were performed at 3 μm resolution, using an Agilent DNA Microarray Scanner, model ‘SYS-SN-ARRAY’. Data analysis was performed using the ADM-2 algorithm, as supplied within ‘Genomic Workbench Standard Edition 5.0’, with routine settings as follows: Aberration Algorithm: ADM-2, Threshold: 6.7, Centralisation: ON, Bin Size: 10, Centralisation Threshold 6.0, Fuzzy Zero: ON, Combine Replicates (Intra Array): OFF, Genome: hg18, Aberration Filters: minProbes=5 AND minAvgAbsLogRatio=0.25 AND maxAberrations=30 AND percent Penetrance=0, Expand Non Unique Probes: OFF. For parental follow-up testing, the Agilent SurePrint G3 Human CGH Microarray 8 × 60K nontargeted array design was used, with effective median resolution of 0.25 Mb. All other test parameters were as above.
PRKD1 and FOXG1 mutation screening
PRKD1 and FOXG1 screening was carried out on 32 atypical Rett syndrome patients, previously excluded for mutations in MECP2 and CDKL5. The coding region of the primary FOXG1 transcript (NM_005249.4), and the 18 exons, including exon–intron boundaries, of PRKD1 (NM_002742.2) were amplified from genomic DNA with primers designed with Primer3 software,23 using GenBank reference sequence NC_000014.8 as the genomic reference. The A in the initiation codon is designated as +1. Primer sequences are given in Supplementary Table 1, with some FOXG1 primers identical to those from other studies.24, 25 Bi-directional sequencing was carried out by Macrogen (Seoul, Korea) using an ABI3730XL sequencing apparatus (Life Technologies, Carlsbad, CA, USA). Sequence traces were analysed using Mutation Surveyor (SoftGenetics, State College, PA, USA).
Expression of FOXG1 and PRKD1 in patients with 14q12 microdeletions
Expression levels of FOXG1 and PRKD1 were measured in a lymphoblastoid cell line derived from white blood cells from case 1 and in whole blood collected in PAXgene Blood RNA tubes (Qiagen, Doncaster VIC, Australia) from cases 2 and 3. Two normal controls for each sample type were available. RNA extractions were carried out using RNeasy Mini Kit (Qiagen) for lymphoblastoid cell lines and PAXgene Blood RNA kit (Qiagen) for blood samples, followed by triplicate cDNA synthesis. Messenger levels for FOXG1, PRKD1 and GAPDH were measured in duplicate using reverse-transcription quantitative PCR using a Corbett Rotorgene 6000 (Qiagen) and associated software. Statistical analysis (nonparametric tests for independent samples) was carried out using IBM SPSS Statistics package, Version 19 (Armonk, NY, USA).
Results
Chromosomal microarray analysis
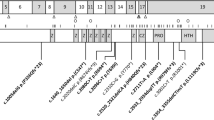
For all three patients in this study, the finding of a chromosome 14q microdeletion was initially determined by clinical comparative genomic hybridisation analysis. Parental samples were also tested, and no parents were found to carry any 14q12 deletion. The three deletions were all within chromosome band 14q12. Further details are outlined in Figure 1. Chromosome coordinates refer to chromosome build HG18.
14q12 microdeletions excluding coding region of FOXG1. Microdeletions at 14q12 were detected in three patients in this report (cases 1–3) by array CGH. The PRKD1 gene was disrupted but FOXG1 remained intact in all three patients. A similar cytogenetic finding has been reported in patient 4 in Kortüm et al.13 There were also two patients of similar phenotypes reported by Shoichet et al31 and Kortüm et al,13 with translocations affecting 14q12 and breakpoints downstream of FOXG1.
The deletion in case 1 was of estimated minimum extent 1.17 Mb, extending from 28.765 to 29.942 Mb. Maximum extent was estimated at 1.21 Mb, from 28.747 to 29.962 Mb. The gene PRKD1 lies within the deleted region. In case 2, the deletion was of minimum size 1.15 Mb, extending from 28.407 to 29.558 Mb (maximum extent was 1.18 Mb, from 28.396 to 29.574). The deletion includes the entire PRKD1 gene, whereas FOXG1 maps 0.1 Mb outside its proximal boundary. In case 3, the minimum deletion size was 0.89 Mb, extending from position 28.499 to 29.386 Mb (maximum size was 0.91 Mb, from 28.487 to 29.393). The deletion includes part of PRKD1, whereas FOXG1 maps 0.18 Mb outside its proximal boundary.
PRKD1 and FOXG1 mutation screening
A single-point mutation of FOXG1 was identified in the cohort of 32 atypical Rett syndrome patients. A cytosine duplication at nucleotide 256 (c.256dupC) was identified in patient 4 with the clinical diagnosis of congenital Rett syndrome variant. The duplication is predicted to lead to a shift in the open-reading frame of the protein (p.Gln86Pro*34). Screening of the parents indicated that this mutation was de novo. The mutation has been previously identified in both a female and a male with congenital variant of Rett syndrome.24, 26
Screening of the same 32 patients did not reveal any pathogenic mutations in PRKD1, a protein kinase involved in extracellular receptor-mediated signal transduction pathways. A number of known nonpathogenic polymorphisms were identified and shown in Table 2. A 5′UTR variation (c.-9C>A) was also identified in one patient. The variation was not reported in the NCBI dbSNP, and parental DNA was not available for screening. The promoter region of PRKD1 was analysed using two software programs (BDGP Neural Network promoter prediction software, http://www.fruitfly.org/seq_tools/promoter.html27 and Promo 3.0, http://alggen.lsi.upc.es/cgi-bin/promo_v3/promo/promoinit.cgi?dirDB=TF_8.3 ).28 There were no differences between the predictions from the wild-type sequence and that containing c.-9C>A. It is unlikely that this sequence variation has any effect on the initiation of transcription of PRKD1. Similarly, prediction for translation initiation using NetStart1.029 (http://www.cbs.dtu.dk/services/NetStart/) gave near-identical results between the wild-type and mutated sequences, suggesting that c.-9C>A also has no effect on translation of PRKD1 protein.
Expression of FOXG1 and PRKD1 in patients with 14q microdeletions
Both FOXG1 and PRKD1 mRNA expression levels appeared to be decreased in whole-blood samples from cases 2 and 3 (Figure 2). Statistical analysis carried out by considering triplicate cDNA as distinct samples, giving n=6, indicated that the expression differences observed in both genes were statistically significantly reduced (nonparametric Mann–Whitney U-test, P<0.005). Using lymphoblast cell lines from case 1 and controls, there was no difference in either FOXG1 or PRKD1 expression between the patient and the controls. However, these triplicates did not represent true biological replicates and inclusion of similar samples in the future would be required to establish the significance of these findings.
Expression of PRKD1 and FOXG1 in 14q12 microdeletion patients. Folds of expression, relative to two normal controls, of FOXG1 (white bars) and PRKD1 (black bars) in the three patients were measured in a lymphoblast cell line for case 1 and in whole blood from cases 2 and 3. Expressions of both genes and a housekeeping gene (glyceraldehyde 3-phosphate dehydrogenase) were measured using triplicate cDNA per sample (means of triplicate cDNA shown, with error bars showing SD).
Discussion
The 14q12 microdeletion is associated with a clinically recognisable phenotype, characterised by severe mental retardation with a normal perinatal period and early delay in achievement of developmental milestones.14 The clinical course resembles Rett syndrome with postnatal microcephaly and growth retardation, hypotonia, epilepsy, stereotypic hand movements and feeding problems. However, unlike classical Rett syndrome, there is early developmental delay with little or no regression and more significant microcephaly. In addition, some patients have been reported to have mild dysmorphism, protruding tongue movements and agenesis of the corpus callosum. The clinical characteristics of patients with mutations of the FOXG1 gene reported to date are similar,13, 30 although there has been one case report of a child with classical Rett syndrome having a FOXG1 mutation.25 There is phenotypic overlap between congenital Rett syndrome variants with FOXG1 mutations and the clinical presentation of our three patients with this 14q12 microdeletion, not encompassing the FOXG1 gene. The commonly deleted region is ∼260 kb and includes the PRKD1 gene but not the FOXG1 gene.
The clinical features of our three cases are consistent with the diagnosis of atypical Rett syndrome of the congenital onset variant.1 Cases 1 and 2 both had abnormal initial development with subsequent severe psychomotor delay, loss of some hand function, midline stereotypic hand movements, postnatal microcephaly, peripheral vasomotor disturbance, breathing abnormalities and tongue stereotypies. The girls did not learn to walk. In addition, they had a seizure disorder. Case 1 had infantile spasms at 1 year of age, whereas case 2 did not have seizures until 11 years of age. Both had agenesis of the corpus callosum on neuroimaging. Case 3 had abnormal initial development with subsequent severe psychomotor delay and some loss of acquired speech, midline stereotypic hand movements, bruxism, cold extremities, tongue stereotypies, impaired sleep pattern and postnatal microcephaly. We believe that there is a subtle pattern of dysmorphology associated with deletions of the 14q12 region. Cases 1 and 2 demonstrate some similarities of facial appearance, including an oval-shaped face, strabismus, full and everted lower lip with a tendency to hold the mouth open. In addition, the females have long fingers and toes, clinodactyly of the second toe (case 1) and shortening of the 4th and 5th metatarsals bilaterally. Case 3 had epicanthic folds. All three cases were described as having a ‘happy disposition’ and tongue thrusting movements.
Other reports of chromosomal abnormalities involving 14q12 have described patients with similar phenotypes to the three cases in this study (summarised in Table 1).7, 8, 12, 14 Of these, there has only been one previous microdeletion of 14q12 not involving FOXG1.13 In addition, there were two translocations affecting chromosome 14 identified in patients with similar phenotypes, although the breakpoints on chromosome 14 mapped 5 and 265 kb downstream of the primary transcript of FOXG1.13, 31 The collective data suggest that cis-regulatory elements exist in the region between FOXG1 and PRKD1, and we propose that they play an important role in the regulation of FOXG1 expression. In silico analysis of the shared-deleted region identified two conserved regions containing potential regulatory elements, located 620 and 684 kb downstream from FOXG1 (see Supplementary Table 2). The former region has also been proposed by Kortüm et al13 to play a regulatory role.13
In support of this hypothesis, a decrease in FOXG1 expression level was indicated in cases 2 and 3, despite its coding region remaining intact in both patients. PRKD1 expression was also affected in both patients, although this might be partially attributed to the disruption of the PRKD1 coding region caused by the microdeletions. Additional information confirming the specificity of the quantitation process can be found in Supplementary Figures S1–S4. The decreases were not observed in case 1, but this may be because of the change in sample type, as a clonal lymphoblastoid cell line was used instead of whole blood for case 1. Gene enhancer assays carried out in transgenic mice by another group indicated that some conserved sequence elements in the deleted region in common may have regulatory roles.13
In most of the reported 14q12 microdeletions thus far, the PRKD1 gene has also been disrupted.8, 12, 13, 14 Including this report, only four cases of 14q12 microdeletions have found FOXG1 gene to be intact, the other being reported by Kortüm et al.13 In earlier reports and mutation analyses, FOXG1 has been considered the primary pathogenic cause in the patients because of its role in the formation of the developing brain.9 FOXG1 is a transcriptional repressor with expression restricted to fetal and adult brain and testis.32 In addition, overlapping expression of FOXG1 and MECP2 has been demonstrated in differentiating forebrain cortex.17 FoxG1 is hypothesised to protect postmitotic neurons from neurotoxic effects of MeCP2.33 Although their exact interaction is yet to be defined, this may suggest a common pathway or interaction at crucial points of brain development.
We propose that the primary defect in our patients is misregulation of the FOXG1 gene rather than a primary abnormality of PRKD1 because of the similarities between the phenotype of those patients with 14q12 microdeletions and those with FOXG1 point mutations, as well as the reductions in FOXG1 expression observed in cases 2 and 3. However, a possible pathogenic role of loss of PRKD1 in all the patients with 14q12 microdeletions cannot be disregarded. PRKD1 encodes a serine–threonine kinase that plays a role in cell proliferation and differentiation, apoptosis, cardiac contraction and cancer.34 There are currently no known mutations or disorders linked to mutations in PRKD1, nor is much known about the mechanism of its regulation of gene expression. We suggest that array CGH is a mandatory investigation in patients with a congenital Rett syndrome phenotype without a pathogenic mutation in MECP2, CDKL5 or FOXG1.
Accession codes
References
Neul JL, Kaufmann WE, Glaze DG et al: Rett syndrome: revised diagnostic criteria and nomenclature. Ann Neurol 2010; 68: 944–950.
Amir RE, Fang P, Yu Z et al: Mutations in exon 1 of MECP2 are a rare cause of Rett syndrome. J Med Genet 2005; 42: e15.
Weaving LS, Christodoulou J, Williamson SL et al: Mutations of CDKL5 cause a severe neurodevelopmental disorder with infantile spasms and mental retardation. Am J Hum Genet 2004; 75: 1079–1093.
White R, Ho G, Schmidt S et al: Cyclin-dependent kinase-like 5 (CDKL5) mutation screening in Rett syndrome and related disorders. Twin Ret Hum Genet 2010; 13: 168–178.
Tao J, Van Esch H, Hagedorn-Greiwe M et al: Mutations in the X-linked cyclin-dependent kinase-like 5 (CDKL5/STK9) gene are associated with severe neurodevelopmental retardation. Am J Hum Genet 2004; 75: 1149–1154.
Rolando S : Rett syndrome: report of eight cases. Brain Dev 1985; 7: 290–296.
Jacob FD, Ramaswamy V, Andersen J, Bolduc FV : Atypical Rett syndrome with selective FOXG1 deletion detected by comparative genomic hybridization: case report and review of literature. Eur J Hum Genet 2009; 17: 1577–1581.
Papa FT, Mencarelli MA, Caselli R et al: A 3 Mb deletion in 14q12 causes severe mental retardation, mild facial dysmorphisms and Rett-like features. Am J Med Genet A 2008; 146A: 1994–1998.
Martynoga B, Morrison H, Price DJ, Mason JO : Foxg1 is required for specification of ventral telencephalon and region-specific regulation of dorsal telencephalic precursor proliferation and apoptosis. Dev Biol 2005; 283: 113–127.
Brancaccio M, Pivetta C, Granzotto M, Filippis C, Mallamaci A : Emx2 and Foxg1 inhibit gliogenesis and promote neuronogenesis. Stem Cells 2010; 28: 1206–1218.
Dou CL, Li S, Lai E : Dual role of brain factor-1 in regulating growth and patterning of the cerebral hemispheres. Cereb Cortex 1999; 9: 543–550.
Bisgaard AM, Kirchhoff M, Tümer Z et al: Additional chromosomal abnormalities in patients with a previously detected abnormal karyotype, mental retardation, and dysmorphic features. Am J Med Genet A 2006; 140: 2180–2187.
Kortüm F, Das S, Flindt M et al: The core FOXG1 syndrome phenotype consists of postnatal microcephaly, severe mental retardation, absent language, dyskinesia, and corpus callosum hypogenesis. J Med Genet 2011; 48: 396–406.
Mencarelli MA, Kleefstra T, Katzaki E et al: 14q12 microdeletion syndrome and congenital variant of Rett syndrome. Eur J Med Genet 2009; 52: 148–152.
Mencarelli MA, Spanhol-Rosseto A, Artuso R et al: Novel FOXG1 mutations associated with congenital variant of Rett syndrome. J Med Genet 2010; 47: 49–53.
Bahi-Buisson N, Nectoux J, Girard B et al: Revisiting the phenotype associated with FOXG1 mutations: two novel cases of congenital Rett variant. Neurogenetics 2010; 11: 241–249.
Ariani F, Hayek G, Rondinella D et al: FOXG1 is responsible for the congenital variant of Rett syndrome. Am J Hum Genet 2008; 83: 89–93.
Brunetti-Pierri N, Paciorkowski AR, Ciccone R et al: Duplications of FOXG1 in 14q12 are associated with developmental epilepsy, mental retardation, and severe speech impairment. Eur J Hum Genet 2011; 19: 102–107.
Striano P, Paravidino R, Sicca F et al: West syndrome associated with 14q12 duplications harboring FOXG1. Neurology 2011; 76: 1600–1602.
Tohyama J, Yamamoto T, Hosoki K et al: West syndrome associated with mosaic duplication of FOXG1 in a patient with maternal uniparental disomy of chromosome 14. Am J Med Genet A 2011; 155A: 2584–2588.
Yeung A, Bruno D, Scheffer IE et al: 4.45 Mb microduplication in chromosome band 14q12 including FOXG1 in a gril with refractory epilepsy and intellectual impairment. Eur J Med Genet 2009; 52: 440–442.
Amor DJ, Burgess T, Tan TY, Pertile MD : Questionable pathogenicity of FOXG1 duplication. Eur J Hum Genet 2012; 20: 595–596.
Rozen S, Slaketsky H : Primer3 on the WWW for general users and for biologist programmers; in Krawetz S, Misener S, (eds): Bioinformatics Methods and Protocols: Methods in Molecular Biology. Totowa, NJ: Humana Press, 2000; Vol 132: pp 365–386.
Le Guen T, Bahi-Buisson N, Nectoux J et al: A FOXG1 mutation in a boy with congenital variant of Rett syndrome. Neurogenetics 2011; 12: 1–8.
Philippe C, Amsallem D, Francannet C et al: Phenotypic variability in Rett syndrome associated with FOXG1 mutations in females. J Med Genet 2010; 47: 59–65.
Takahashi S, Matsumoto N, Okayama A et al: FOXG1 mutations in Japanese patients with the congenital variant of Rett syndrome. Clin Genet 2011, e-pub ahead of print 1 December 2011 doi:10.1111/j.1399-0004.2011.01819.x.
Reese MG : Application of a time-delay neural network to promoter annotation in the Drosophila melanogaster genome. Comput Chem 2001; 26: 51–56.
Messeguer X, Escudero R, Farré D, Nuñez O, Martínez J, Albà M : PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics 2002; 18: 333–334.
Pedersen AG, Nielsen H : Neural network prediction of translation initiation sites in eukaryotes: perspectives for EST and genome analysis. Proc Int Conf Intell Syst Mol Biol 1997; 5: 226–233.
Sampieri K, Meloni I, Scala E et al: Italian Rett database and biobank. Hum Mutat 2007; 28: 329–335.
Shoichet SA, Kunde SA, Viertel P et al: Haploinsufficiency of novel FOXG1B variants in a patient with severe mental retardation, brain malformations and microcephaly. Hum Genet 2005; 117: 536–544.
Murphy DB, Wiese S, Burfeind P et al: Human brain factor 1, a new member of the fork head gene family. Genomics 1994; 21: 551–557.
Dastidar SG, Bardai FH, Ma C et al: Isoform-specific toxicity of Mecp2 in postmitotic neurons: suppression of neurotoxicity by FoxG1. J Neurosci 2012; 32: 2846–2855.
Steinberg SF : Regulation of protein kinase D1 activity. Mol Pharmacol 2012; 81: 284–291.
Le Guen T, Fichou Y, Nectoux J et al: A missense mutation within the fork-head domain of the Forkhead Box G1 gene (FOXG1) affects tis nuclear localization. Hum Mutat 2010; 32: E2026–E2035.
Van der Aa N, Van den Bergh M, Ponomarenko N, Verstraete L, Ceulemans B, Storm K : Analysis of FOXG1 is highly recommended in male and female patients with Rett syndrome. Mol Syndromol 2011; 1: 290–293.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Ellaway, C., Ho, G., Bettella, E. et al. 14q12 microdeletions excluding FOXG1 give rise to a congenital variant Rett syndrome-like phenotype. Eur J Hum Genet 21, 522–527 (2013). https://doi.org/10.1038/ejhg.2012.208
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2012.208
Keywords
This article is cited by
-
Hypoventilation and sleep hypercapnia in a case of congenital variant-like Rett syndrome
Italian Journal of Pediatrics (2022)
-
Diagnosis of FOXG1 syndrome caused by recurrent balanced chromosomal rearrangements: case study and literature review
Molecular Cytogenetics (2020)
-
A novel 14q13.1–21.1 deletion identified by CNV-Seq in a patient with brain-lung-thyroid syndrome, tooth agenesis and immunodeficiency
Molecular Cytogenetics (2019)
-
Regulatory variants of FOXG1 in the context of its topological domain organisation
European Journal of Human Genetics (2018)
-
Partial monosomy14q involving FOXG1 and NOVA1 in an infant with microcephaly, seizures and severe developmental delay
Molecular Cytogenetics (2016)