Abstract
Treacher–Collins–Franceschetti syndrome (TCS) is an autosomal dominant craniofacial disorder characterised by midface hypoplasia, micrognathia, downslanting palpebral fissures, eyelid colobomata, and ear deformities that often lead to conductive deafness. A total of 182 patients with signs consistent with a diagnosis of TCS were screened by DNA sequence and dosage analysis of the TCOF1 gene. In all, 92 cases were found to have a pathogenic mutation by sequencing and 5 to have a partial gene deletion. A further case had a novel in-frame deletion in the alternatively spliced exon 6A of uncertain pathogenicity. The majority of the pathogenic sequence changes were found to predict premature protein termination, however, four novel missense changes in the LIS1 homology motif at the 5′ end of the gene were identified. The partial gene deletions of different sizes represent ∼5.2% of all the pathogenic TCOF1 mutations identified, indicating that gene rearrangements account for a significant proportion of TCS cases. This is the first report of gene rearrangements resulting in TCS. These findings expand the TCOF1 mutation spectrum indicating that dosage analysis should be performed together with sequence analysis, a strategy that is predicted to have a sensitivity of 71% for patients in whom TCS is strongly suspected.
Similar content being viewed by others
Introduction
Treacher–Collins–Franceschetti syndrome (TCS) is a rare autosomal dominant congenital disorder of craniofacial morphogenesis that occurs with an estimated prevalence of 1/50 000 live births. The syndrome is characterised by bilaterally symmetrical craniofacial abnormalities. The prominent clinical features are malar hypoplasia caused by hypoplasia of the zygomatic complex, mandibular hypoplasia, downslanting palpebral fissures, colobomata of the lower eyelids, and ear malformations often associated with bilateral conductive hearing loss. TCS has extreme inter- and intra-familial phenotypic variability, which ranges from perinatal death owing to a compromised airway to a phenotype that goes undetected by medical examination.1 Penetrance is thought to be high but non-penetrance has been reported.2
Deleterious mutations in TCOF1 (OMIM *606847) are associated with TCS type 1 (OMIM #154500).3 The longest transcript of TCOF1 comprises 26 exons, which include the alternatively spliced exons 6A and 19.4 Other minor splice isoforms include transcripts lacking exon 6A, transcripts lacking exon 19 and transcripts that include a novel exon 16A.4 TCOF1 encodes a low complexity, serine/alanine-rich protein called treacle, which has unique N and C termini and a large central repeat domain containing motifs shared with nucleolar trafficking proteins.5 Treacle is therefore thought to be a nucleolar phosphoprotein involved in ribosomal DNA gene transcription.6 The C-terminal region of treacle is important for localisation to the nucleolus 7, 8 and the N-terminal region contains a LIS1 homology motif that may contribute to the regulation of microtubule dynamics, either by mediating dimerisation, or else by binding cytoplasmic dynein heavy chain or microtubules directly.9 As the structures affected in TCS patients arise from the first and second branchial arches during early embryogenesis,10 the abnormal development due to treacle haploinsuffiency may be caused by inhibition of production of properly modified mature ribosomal RNA (rRNA) as well as inhibition of rRNA gene transcription in the prefusion neural folds during the early stages of embryogenesis, consequently affecting proliferation and proper differentiation of these embryonic cells.11 The recent identification of mutations in genes encoding subunits of RNA polymerases I and III (POLR1D and POLR1C) in a small subset of patients with TCS, confirm that TCS is a ribosomopathy and genetically heterogeneous.12
Mutations have been identified throughout the TCOF1 gene and the vast majority identified to date are unique to the family studied and introduce a premature termination codon; missense mutations are rare and confined to the 5′ end of the gene (http://genoma.ib.usp.br/TCOF1_database and references therein). The literature suggests that 62–82% of patients diagnosed with TCS have the diagnosis confirmed by sequence analysis of TCOF1, POLR1D or POLR1C,1, 2, 12, 13 however, the aetiology of the remainder remains uncertain. Gross deletions or other rearrangements of TCOF1 cannot be ruled out as they have only been investigated in one study by Splendore et al,14 who found no alterations in seven patients. This study looks at a larger cohort of 182 subjects using sequencing and MLPA.
Patients and methods
Patients
The cohort of patients studied included 182 unrelated individuals with clinical features of TCS referred to the Oxford Regional Clinical Molecular Genetics laboratory between May 2005 and June 2011. A review of the referral information by an experienced dysmorphologist indicated that 119 of these cases were strongly suspected to have TCS, whereas 55 cases had a less secure diagnosis. In eight cases insufficient clinical information was available to make an assessment (Table 1). Patients were sequenced initially, and dosage analysis then carried out on those cases in whom no definitely pathogenic mutation could be found. Where available, parents were also analysed to determine if variants had arisen de novo.
PCR and sequencing
The 27 coding exons (including exons 6A, 16A and 19)4 were amplified in 28 fragments (primer sequences and reaction conditions given in Supplementary Table 1). PCRs were purified using paramagnetic beads (Agencourt: Beckman Coulter plc, High Wycombe, UK), then sequenced using BigDye v3.1 chemistry (Applied Biosystems, Life Technologies Corporation, Carlsbad, CA, USA). Fragments were separated and analysed on an ABI3730 using Sequencing Analysis 5.2 software (Applied Biosystems). Mutation Surveyor software (SoftGenetics, State College, Philadelphia, PA, USA) was used to identify sequence variants, and in silico splice prediction software used to predict alterations to splice sites (Alamut version 1.5 or 2.0, Interactive Biosoftware, Rouen, France). Nomenclature for sequence variants is in accordance with HGVS guidelines (www.hgvs.org) using GenBank NCBI reference sequence NM_001135243.1 (this includes exon 6A, but excludes exon 16A), as recommended by Splendore et al.14
MLPA
The P310-A1 or P310-B1 TCOF MLPA kit (MRC-Holland) was used according to the manufacturer's recommendations, and the reactions analysed using a 3730 automated analyser and GeneMapper software (Applied Biosystems). Statistical analysis of the data was carried out using a MLPA data analysis spreadsheet devised by the National Genetics Reference Laboratory, Manchester, UK (http://www.ngrl.org.uk/Manchester), with results presented as a bar chart. The P310-A1 and P310-B1 kits target exons 1–7, 9–18 (including exon 6A) and 21–26 (exon 26 is untranslated). Although several probes were designed for exons 19 and 20 none were robust enough for inclusion in these kits, however, there are only 2.8 kb between the flanking probes in exons 18 and 21. Exon 16A is also not targeted directly, but a probe-pair hybridises to sequence 760 nucleotides after the last base of this exon. The P310-A1 kit also contained a probe pair targeting exon 8, but due to problems with reliability it has been removed from the later B1 version of the kit.
Quantitative PCR
Quantitative PCR (qPCR) was performed using the SYBR Green detection method and an ABI Prism 7500 Sequence Detection System (Applied Biosystems). All amplicons were 100–120 nucleotides in length (primer sequences given in Supplementary Table 1). Reactions were amplified in 20 μl volumes using 1x SYBR Green mastermix (Applied Biosystems), 0.2 μ M of each primer and 10 ng genomic DNA. PCR conditions were as follows: 2 min initial step of 50 °C then denaturation for 10 min at 95 °C followed by 40 cycles of 95 °C for 15 sec and 60 °C for 1 min, ending with a final dissociation. The Sequence Detection System software (SDS version 1.2; Applied Biosystems) was used to analyse the qPCR data. The copy number was estimated using the comparative ΔΔCt method where a normal control has a ratio of one.15
Array CGH (aCGH)
The TCOF1 gene region was targeted using a custom 180K oligoarray (Agilent Technologies, Wokingham, UK. Details on request). The array includes four probes in TCOF1 as well as probes in surrounding genes, including one in CD74. Analysis was carried out on a G2505B/G2539A microarray scanner (Agilent Technologies) that scans at 2 μm resolution.
Results
Sequencing analysis
A total of 79 different pathogenic sequence variants (60 novel) were identified in 92 unrelated individuals, spread throughout the gene (Table 2). Of these, 59 are predicted to disrupt protein translation by introducing premature stop codons, or by abolishing the translation-start site. Four further missense changes in exon 1 (p.(Ile14Met), p.(His17Arg), p.(Ala21Pro), p.(Ala27Thr)) alter highly conserved amino acids in the LIS1 homology domain. In addition, two of these missense variants have arisen de novo. Fourteen further variants were predicted to disrupt splicing. Two variants, c.4366_4369delGAAA and c.4362_4365delAAAA, predict a frameshift that results in the replacement of the final 33 amino acids with 117 missense amino acids. No pathogenic sequence variants were detected in the alternatively spliced exon 16A; however, an in-frame novel deletion of 18 nucleotides (c.827_844del, p.(Gly276_Glu281del)) was detected in exon 6A in a case with extensive family history. The pathogenicity of this variant is currently uncertain.
A total of 27 other variants (20 novel) were identified during the screening of the cohort (Table 3). One of the novel variants could be classified as a non-pathogenic single-nucleotide polymorphism (SNP) on the basis that it was seen in multiple individuals. Five further unvalidated variants already recorded in dbSNP could be assigned as non-pathogenic from the data (Table 3). The other novel variants, however, could not be definitively classified as non-pathogenic SNPs, but were considered unlikely to be pathogenic. This was supported by parental studies in five cases.
MLPA analysis
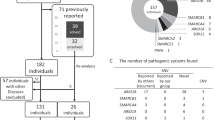
MLPA analysis identified partial gene deletions of TCOF1 in five patients (Figure 1). Patients 170 and 96 were shown to have deletion of a single MLPA probe, located in exons 1 and 26 (which encodes 3′-UTR), respectively. Reduced hybridisation due to SNPs under the probe-binding sites was ruled out by sequencing. Validation of these results for patients 170 and 96 was carried out by qPCR (see below). The three other patients showed reduced hybridisation to multiple adjacent probes consistent with a contiguous deletion of exons 1–15 (patient 452), exons 23–26 (patient 8) and exons 1–6 (patient 226).
Dosage results for the five patients with a partial TCOF1 deletion. A 50% relative bar height indicates a heterozygous deletion. MLPA detected a heterozygous deletion of: (452) exons 1–15; (170) exon 1; (96) exon 26; (8) exons 23–26; (226) exons 1–6. Grey bars on the left represent control probes, white bars represent probes located within or nearby to all TCOF1-coding exons except exons 19 and 20. The grey bar on the right represents the untranslated TCOF1 exon 26 (3′UTR).
Quantitative PCR
For patients 170 and 96, qPCRs were carried out to determine the extent of the deletion and to provide confirmation of the MLPA result (Figure 2). In-patient 170, data showed that the deletion encompassed the translation initiation signal, as well as exon 1 coding sequence. The deletion in patient 96 was shown to extend beyond the validated polyadenylation signal within exon 26,16 removing a large part of the 3′-UTR. However, the deletion did not appear to encompass the intron 25 splice acceptor site.
Schematic illustrating results of MLPA and quantitative PCR analysis of TCOF1 in (a) 5′-UTR region in case 170 and (b) 3′-UTR region in case 96. Coding sequence (black boxes), UTR (hatched boxes) and introns (thin black lines). Black lines above the figure indicate the target sites for the MLPA probes and the amplicons for qPCR relative copy number calculated from the quantitative PCR assay in the respective patient and a deletion control are shown in the graphs above each amplicon. ATG is the translation initiation signal, TGA is the translation termination signal and AATAAA is the polyadenylation signal. The dark grey line below the figure indicates deleted sequence where probes/amplicons show 50% dosage; the light grey line indicates sequence that is not deleted where probes/amplicons show normal dosage. Grey dashed lines show regions of uncertainty. Not to scale.
aCGH
Karyotyping and aCGH analysis were carried out on the affected brother of patient 8, in whom the exon 23–26 deletion was also demonstrated by MLPA. The deletion was not visible by karyotyping. Of the aCGH probes, only one is known to be within the deletion as it lies in intron 25. Results for this probe were consistent with it being deleted. There was no evidence for deletion of adjacent probes, indicating that the deletion does not extend into the adjacent gene CD74.
Discussion
Of the 182 unrelated diagnostic referrals, 92 were found to have a pathogenic sequence variant and 5 a pathogenic gene rearrangement (Table 1). These are the first gene rearrangements described in TCOF1. The five deletions constitute 5.2% of all the pathogenic variants that were detected in TCOF1 in this study. Of the patients with sequence changes, 57% had a frameshifting small deletion or insertion, or a mutation disrupting the start codon, 23% had a nonsense mutation, 16% had a variant predicted to affect splicing, and 4% had a pathogenic missense mutation (Table 4). This spectrum is similar to that described in previous studies that have shown that the majority of mutations in TCS1 lead to loss of protein function consistent with a mechanism of haploinsufficiency (http://genoma.ib.usp.br/TCOF1_database and references therein). The vast majority of mutations identified were only observed in one family consistent with previous studies that have shown that the majority of disease-causing mutations in TCOF1 are private. The exceptions were c.304+5G>C, c.3163C>T (p.(Gln1055*)), c.4218dupG (p.(Ser1407fs)), and c.4365delA (p.(Glu1456fs)), each found in two patients, and c.4369_4373delAAGAA (p.(Lys1457fs)) found 10 times. This last deletion has been reported several times previously (Table 1) and occurs within a repeat motif making it a mutational hotspot within the gene. Consistent with this hypothesis was the finding that this deletion had arisen de novo in two of the cases. Stratifying the mutation data by clinical phenotype (Table 1) indicated that overall pathogenic point mutations were identified in 51% of all referrals; this was increased to 68% if only those cases with a high clinical suspicion of TCS were included. Including MLPA data as well increased sensitivity to 71% in this group.
Of the missense variants that were identified four could be categorised as being likely to be pathogenic based on their location at highly conserved residues within the LIS1 homology domain, where other pathogenic missense changes have been reported.1, 13, 17, 18, 19 Patient 443, who presented with micrognathia, deficient zygomatic arches, minor downslant of palpebral fissures, only rudimentary condyles, but no obvious hearing loss, was found to have c.42C>G (p.(Ile14Met)). Another variant at this residue, p.(Ile14Phe), has been reported previously in association with TCS.18 The equivalent residue to p.Ile14 in the Lis1 protein is important in forming hydrophobic contacts between Lis1 homodimers.20 Parental analysis indicated that the p.(Ile14Met) variant had been inherited from the clinically unaffected father, however, such phenotypic variability of mutations, even within families has been reported previously.2 Patient 67, who has a clinical diagnosis of TCS, was found to have c.50A>G (p.(His17Arg)). The equivalent residue in the Lis1 protein is important for the forming of both hydrophobic contacts and hydrogen bonds between Lis1 homodimers,20 and analysis of parental samples indicated that the mutation had arisen de novo. Patient 125, who had features suggestive of TCS, was found to have c.61G>C (p.(Ala21Pro)). Parental analysis indicated that it had arisen de novo. Finally, Patient 270, who had a clinical diagnosis of TCS, was found to have c.79G>A (p.(Ala27Thr)). Another variant at the preceding residue c.77C>T (p.(Ala26Val)) has been described previously and was apparently de novo.19 Parental samples were not available to determine if the newly identified variant was de novo.
A further variant, that was initially considered to be a missense change (c.3613G>A, p.(Gly1205Ser)), was classified as being likely to be pathogenic due to in silico predictions of aberrant splicing, combined with literature evidence. Patient 110 presented with symptoms suggestive of TCS comprising bilateral dysplastic ears and abnormal lower eyelids with sparse lower lid eyelashes. Clinical observations did not identify malar hypoplasia, and hearing is normal. A sister is also affected. The exon 22 variant is predicted to alter a residue conserved across many vertebrate species, but is in the central repeat region of the protein where pathogenic missense changes have not been reported. However, a de novo variant at an adjacent nucleotide (c.3612A>C) has been reported21 that caused precise skipping of exon 22, causing a frameshift and predicted premature termination of translation. In silico analysis showed that the aberrant splicing could be due to disruption of an exonic splice enhancer.21 Analysis using splice site prediction software suggested that the c.3613G>A variant in patient 110 might also disrupt this exonic splice enhancer and therefore could also lead to aberrant splicing of exon 22. Unfortunately, an RNA sample was not available to test this experimentally. Family studies indicated that the variant was also carried by the affected sister and had been inherited from the clinically unaffected mother.
Two further exonic variants were also considered likely to affect splicing. The first variant (c.3183G>A) affects the last base of exon 18. Substitution of this G with a T nucleotide has been reported to cause skipping of exons 18 to 19 in another patient,22 and analysis using splice-site predictor software indicates that the G>A substitution would also reduce the splicing efficiency of the intron 18 splice donor site. The second variant (c.3156C>T), also within exon 18, is a synonymous change. However, splice prediction software indicates that it may create a cryptic splice donor site 29 bp upstream of the actual intron 19 splice donor site. Unfortunately, RNA samples were not available to test these hypotheses.
An intronic variant, c.566–10C>A, was also identified in patient 631 that was considered likely to be pathogenic. In silico splice-prediction software indicated that this variant might abolish the usual intron 5 splice-acceptor site and replace it with a novel, cryptic site 8 nucleotides upstream. If used, this would lead to the inclusion of 8 nucleotides from intron 5 into the mRNA resulting in a translational frameshift. Although RNA and parental samples were not available to test this hypothesis a similar mechanism was proposed for another variant, c.2196G>A, that was confirmed by RT-PCR analysis.23
Two other novel variants identified were considered less likely to be pathogenic, but could not be ruled out entirely as no other obviously pathogenic change could be identified. In the first case, patient 195 presented with microtia, atresia of the external ear canal, mild mandibular hypoplasia and a bifid uvula and was found to have c.2762C>T (p.(Pro921Leu)) in exon 16 of TCOF1. Pro921 is not highly conserved between vertebrate species and lies within the central repeat region of the protein, where missense mutations have not been reported. Splice-prediction software did not predict any effect on splicing, but without experimental evidence this cannot be ruled out. The second case, patient 18, was found to have a maternally inherited in-frame deletion, c.4377_4379delGAA (p.(Lys1460del)), in exon 24 of TCOF1 within a lysine-rich region near the C-terminus of Treacle that is not highly conserved. This variant has been found in another TCS patient (TCOF1 Mutation Database, http://genoma.ib.usp.br/TCOF1_database/) as have other similar variants considered non-pathogenic; p.(Lys1455_Lys1457del), found in a patient with another de novo pathogenic mutation,19 and p.(Lys1460dup), also found in a control sample.2
Of the patients with deletions, all had a clinical diagnosis of TCS. The full extent of each deletion is not known as they involved either the first or last exons and therefore it cannot be ruled out that the deletions extend beyond the TCOF1 gene into other nearby genes. TCOF1 maps to 5q32 in a gene dense region; 10 genes map within 0.5 Mb 5′ of TCOF1 and 9 genes map within 0.5 Mb 3′ of TCOF1 (http://www.ensembl.org/).
In three cases the deletions identified could extend beyond the 5′-UTR into upstream genes. In-patient 170 the deletion in exon 1 was shown to encompass the translation initiation signal and at least part of the 5′-UTR. She presented as an isolated case diagnosed clinically, however, parental samples were not available to determine whether this is a de novo rearrangement. Patient 452, who has a deletion encompassing exons 1–15, has mandibular hypoplasia, cleft palate, down-slanting palpebral fissures, wide set eyes and abnormal palmar creases. Testing of the parents showed that the deletion is apparently de novo. Patient 226 has a deletion encompassing exons 1–6. He presented with a clinical diagnosis of TCS and has an affected son who also carries the deletion.
Of the nearest upstream genes, CDX1 has been shown to have a phenotypic effect when the orthologous gene is knocked out in mice. CDX1 lies ∼173 kb upstream of TCOF1 and encodes caudal-type homeobox transcription factor 1. The caudal-type transcription factors interact with HOX genes to generate the anterior–posterior axis, and viable homozygous cdx1 knockout mice were shown to have anterior homeotic transformations of vertebrae concomitant with posterior shifts of hox gene expression in the somatic mesoderm.24 The effect of haploinsufficiency of this gene in humans is not known but altered precise temporal and spatial expression of genes such as the HOX genes during embryonic development is required for accurate skeletal modelling.25 Unfortunately, material was not available for aCGH to identify the true extent of the deletions beyond the first exon in these patients.
In the other two cases the deletion extended into the 3′-UTR, and may therefore extend into downstream genes. Patient 8 presented with dysmorphic features consistent with a diagnosis of TCS, and was shown to have a deletion of exons 23–26, that removes the C-terminal region of treacle important for localisation to the nucleolus. Array CGH analysis suggested that the deletion did not extend into the downstream CD74 gene. His brother and mother were shown to be heterozygous for the same deletion, but neither individual expresses the characteristic phenotypic features associated with TCS. Such phenotypic variability has also been reported with TCOF1 point mutations.1, 2 The deletion in patient 96, who presented with mild TCS, and his more severely affected brother, does not encompass any coding sequence but removes part of the 3′-UTR that contains the polyadenylation signal for the major transcript. This is a sequence motif recognised by RNA-binding factors and is essential for transcriptional termination and efficient polyadenylation of mRNAs and release of the polyadenylated mRNA for export from the nucleus.26 Mutations affecting a polyadenylation signal and the secondary structure of the 3′-UTR of mRNA have been shown to cause translation de-regulation27 and have been reported in other diseases.28, 29, 30, 31 It is therefore predicted that deletion of this signal in TCOF1 would lead to reduced accumulation of treacle, consistent with a mechanism of haploinsufficiency in TCS. Parental samples were not available.
The two closest distal genes to TCOF1: CD74 and RPS14, lie ∼2 and 43 kb downstream from TCOF1, respectively. CD74 encodes the major histocompatibility complex II-associated invariant chain (CD74) that functions as an MHC class II chaperone and is required for B-cell development in mice.32 RPS14 encodes the ribosomal protein S14. In humans, heterozygous somatic deletions of RPS14 are thought to be the major cause of 5q- syndrome;33 an acquired myelodysplastic syndrome subtype, characterized by a defect in erythroid differentiation.
No pathogenic TCOF1 gene alterations were identified in 84 remaining patients in the cohort (Table 1). It is possible that pathogenic variants in introns and regulatory regions not covered by the sequencing assay, or deletions and inversions undetectable by sequencing and MLPA may exist in TCOF1 in these patients, particularly those 34 with signs strongly suggestive of TCS. The MLPA kits P310-A1 and P310-B1 (MRC-Holland) that were used do not contain probes for exons 19 or 20, however, while there remains the possibility that patients in this cohort have an undetected deletion or duplication of either or both of these exons, the chance of this is low as the exons are within a short 2.8-kb region between the adjacent probes in exons 17 and 21. It is also possible that mutations in one or more other genes are responsible for the TCS phenotype in these patients, such as the POLR1D and POLR1C genes recently reported.12
In conclusion, these findings suggest that gene rearrangements are responsible for a significant proportion (5.2%) of TCS-associated mutations. The proximity of nearby genes likely to have a phenotypic effect if haploinsufficient predicts that any deletions in patients without unusual additional symptoms would be submicroscopic. It is therefore recommended that dosage analysis by MLPA or a comparable method should be undertaken as part of a TCOF1 gene screening service. These data suggest that combined sequencing and dosage analysis using MLPA has a test sensitivity of around 71% for patients that are referred because TCS is strongly suspected.
References
Splendore A, Jabs EW, Passos-Bueno MR : Screening of TCOF1 in patients from different populations: confirmation of mutational hotspots and identification of a novel missense mutation that suggests an important functional domain in the protein treacle. J Med Genet 2002; 39: 493–495.
Teber OA, Gillessen-Kaesbach G, Fischer S et al: Genotyping in 46 patients with tentative diagnosis of Treacher Collins syndrome revealed unexpected phenotypic variation. Eur J Hum Genet 2004; 12: 879–890.
The Treacher Collins Syndrome Collaborative Group, Dixon J, Edwards SJ et al: Positional cloning of a gene involved in the pathogenesis of Treacher Collins syndrome. Nat Genet 1996; 12: 130–136.
So RB, Gonzales B, Henning D, Dixon J, Dixon MJ, Valdez BC : Another face of the Treacher Collins syndrome (TCOF1) gene: identification of additional exons. Gene 2004; 328: 49–57.
Wise CA, Chiang LC, Paznekas WA et al: TCOF1 gene encodes a putative nucleolar phosphoprotein that exhibits mutations in Treacher Collins Syndrome throughout its coding region. Proc Natl Acad Sci USA 1997; 94: 3110–3115.
Valdez BC, Henning D, So RB, Dixon J, Dixon MJ : The Treacher Collins syndrome (TCOF1) gene product is involved in ribosomal DNA gene transcription by interacting with upstream binding factor. Proc Natl Acad Sci USA 2004; 101: 10709–10714.
Marsh KL, Dixon J, Dixon MJ : Mutations in the Treacher Collins syndrome gene lead to mislocalization of the nucleolar protein treacle. Hum Mol Genet 1998; 7: 1795–1800.
Winokur ST, Shiang R : The Treacher Collins syndrome (TCOF1) gene product, treacle, is targeted to the nucleolus by signals in its C-terminus. Hum Mol Genet 1998; 7: 1947–1952.
Emes RD, Ponting CP : A new sequence motif linking lissencephaly, Treacher Collins and oral-facial-digital type 1 syndromes, microtubule dynamics and cell migration. Hum Mol Genet 2001; 10: 2813–2820.
Sulik KK, Johnston MC, Smiley SJ, Speight HS, Jarvis BE : Mandibulofacial dysostosis (Treacher Collins syndrome): a new proposal for its pathogenesis. Am J Med Genet 1987; 27: 359–372.
Gonzalez B, Henning D, So RB, Dixon J, Dixon MJ, Valdez BC : The Treacher Collins syndrome (TCOF1) gene product is involved in pre-rRNA methylation. Hum Mol Genet 2005; 14: 2035–2043.
Dauwerse JG, Dixon J, Seland S et al: Mutations in genes encoding subunits of RNA polymerases I and III cause Treacher Collins syndrome. Nat Genet 2011; 43: 20–22.
Edwards SJ, Gladwin AJ, Dixon MJ : The mutational spectrum in Treacher Collins syndrome reveals a predominance of mutations that create a premature termination codon. Am J Hum Genet 1997; 60: 515–524.
Splendore A, Fanganiello RD, Masotti C, Morganti LSC, Passos-Bueno MR : TCOF1 mutation database: Novel mutation in the alternatively spliced exon 6A and update in mutation nomenclature. Hum Mutat 2005; 25: 429–434.
Livak KJ, Schmittgen TD : Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method. Methods 2001; 25: 402–408.
Dixon J, Edwards SJ, Anderson I, Brass A, Scambler PJ, Dixon MJ : Identification of the complete coding sequence and genomic organization of the Treacher Collins syndrome gene. Genome Res 1997; 7: 223–234.
Ellis PE, Dawson M, Dixon MJ : Mutation testing in Treacher Collins syndrome. J Orthod 2002; 29: 293–297.
Dixon J, Ellis I, Bottani A, Temple K, Dixon MJ : Identification of mutations in TCOF1: use of molecular analysis in the pre- and postnatal diagnosis of Treacher Collins syndrome. Am J Med Genet 2004; 127: 244–248.
Fujioka H, Ariga T, Horiuchi K et al: Detection of a novel silent deletion, a missense mutation and a nonsense mutation in TCOF1. Pediatr Int 2008; 50: 806–809.
Kim MH, Cooper DR, Oleksy A et al: The structure of the N-terminal domain of the product of the lissencephaly gene Lis1 and its functional implications. Structure 2004; 12: 987–998.
Macaya D, Katsanis SH, Hefferon TW et al: A synonymous mutation in TCOF1 causes Treacher Collins syndrome due to mis-splicing of a constitutive exon. Am J Med Genet 2009; 149: 1624–1627.
Horiuchi K, Ariga T, Fujioka H et al: Mutational analysis of the TCOF1 gene in 11 Japanese patients with Treacher Collins Syndrome and mechanism of mutagenesis. Am J Med Genet 2005; 134: 363–367.
Gladwin AJ, Dixon J, Loftus SK et al: Treacher Collins syndrome may result from insertions, deletions or splicing mutations, which introduce a termination codon into the gene. Hum Mol Genet 1996; 5: 1533–1538.
Subramanian V, Meyer BI, Gruss P : Disruption of the murine homeobox gene Cdx1 affects axial skeletal identities by altering the mesodermal expression domains of Hox genes. Cell 1995; 83: 641–653.
Burke AC, Nelson CE, Morgan BA, Tabin C : Hox genes and the evolution of vertebrate axial morphology. Development 1995; 121: 333–346.
Danckwardt S, Hentze MW, Kulozik AE : 3′ end mRNA processing: molecular mechanisms and implications for health and disease. EMBO J 2008; 27: 482–498.
Chatterjee S, Pal JK : Role of 5′- and 3′-untranslated regions of mRNAs in human diseases. Biol Cell 2009; 101: 251–262.
Losekoot M, Fodde R, Harteveld CL et al: Homozygous beta+ thalassaemia owing to a mutation in the cleavage-polyadenylation sequence of the human beta globin gene. J Med Genet 1991; 28: 252–255.
Harteveld CL, Losekoot M, Haak H, Heister GA, Giordano PC, Bernini LF : A novel polyadenylation signal mutation in the α2-globin gene causing α-thalassaemia. Br J Haematol 1994; 87: 139–143.
Prior JF, Lim E, Lingam N, Raven JL, Finlayson J : A moderately severe α-thalassemia condition resulting from a combination of the α2 polyadenylation signal (AATAAA → AATA--) mutation and a 3.7 Kb α gene deletion in an Australian family. Hemoglobin 2007; 31: 173–177.
Bennett CL, Brunkow ME, Ramsdell F et al: A rare polyadenylation signal mutation of the FOXP3 gene (AAUAAA → AAUGAA) leads to the IPEX syndrome. Immunogenetics 2001; 53: 435–439.
Shachar I, Flavell RA : Requirement for invariant chain in B cell maturation and function. Science 1996; 274: 106–108.
Ebert BL, Pretz J, Bosco J et al: Identification of RPS14 as a 5q- syndrome gene by RNA interference screen. Nature 2008; 451: 335–339.
Shoo BA, McPherson E, Jabs EW : Mosaicism of a TCOF1 mutation in an individual clinically unaffected with Treacher Collins syndrome. Am J Med Genet A 2004; 126A: 84–88.
Splendore A, Silva EO, Alonso LG et al: High mutation detection rate in TCOF1 among Treacher Collins syndrome patients reveals clustering of mutations and 16 novel pathogenic changes. Hum Mutat 2000; 16: 315–322.
Horiuchi K, Ariga T, Fujioka H et al: Treacher Collins syndrome with craniosynostosis, choanal atresia, and esophageal regurgitation caused by a novel nonsense mutation in TCOF1. Am J Med Genet 2004; 128A: 173–175.
Acknowledgements
We would like to thank all the clinicians who have referred patients to our service for molecular genetic analysis; our colleagues in the Regional Cytogenetics laboratory, Churchill Hospital, Oxford for sharing results of karyotyping analysis, and for assistance with the design and analysis of quantitative PCR assays; and Professor Andrew Wilkie for classifying TCS referrals into those with a strong clinical suspicion and those without, and for critical reviewing of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Bowman, M., Oldridge, M., Archer, C. et al. Gross deletions in TCOF1 are a cause of Treacher–Collins–Franceschetti syndrome. Eur J Hum Genet 20, 769–777 (2012). https://doi.org/10.1038/ejhg.2012.2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2012.2
Keywords
This article is cited by
-
Identification of three novel TCOF1 mutations in patients with Treacher Collins Syndrome
Human Genome Variation (2021)
-
TCOF1 pathogenic variants identified by Whole-exome sequencing in Chinese Treacher Collins syndrome families and hearing rehabilitation effect
Orphanet Journal of Rare Diseases (2019)
-
Mutation screening of Chinese Treacher Collins syndrome patients identified novel TCOF1 mutations
Molecular Genetics and Genomics (2018)
-
Large deletions encompassing the TCOF1 and CAMK2A genes are responsible for Treacher Collins syndrome with intellectual disability
European Journal of Human Genetics (2014)
-
Treacher Collins syndrome: clinical implications for the paediatrician—a new mutation in a severely affected newborn and comparison with three further patients with the same mutation, and review of the literature
European Journal of Pediatrics (2012)