Abstract
Retinoblastoma (Rb) results from inactivation of both alleles of the RB1 gene located in 13q14.2. Whole-germline monoallelic deletions of the RB1 gene (6% of RB1 mutational spectrum) sometimes cause a variable degree of psychomotor delay and several dysmorphic abnormalities. Breakpoints in 12 Rb patients with or without psychomotor delay were mapped to specifically define the role of chromosomal regions adjacent to RB1 in psychomotor delay. A high-resolution CGH array focusing on RB1 and its flanking region was designed to precisely map the deletion. Comparative analysis detected a 4-Mb critical interval, including a candidate gene protocadherin 8 (PCDH8). PCDH8 is thought to function in signalling pathways and cell adhesion in a central nervous system-specific manner, making loss of PCDH8 one of the probable causes of psychomotor delay in RB1-deleted patients. Consequently, we propose to systematically use high-resolution CGH in cases of partial or complete RB1 deletion encompassing the telomeric flanking region to characterize the putative loss of PCDH8 and to better define genotype/phenotype correlations, eventually leading to optimized genetic counselling and psychomotor follow-up.
Similar content being viewed by others
Introduction
Retinoblastoma (Rb) is a rare embryonic neoplasm of retinal origin resulting from inactivation of both alleles of the RB1 gene (MIM no.180200) located in chromosome band 13q14.2.1 Predisposition to Rb must be suspected independently of family history and regardless of the clinical presentation, because 100 and 10% of bilateral and unilateral cases, respectively, carry an inherited or de novo germline mutation. Screening for the predisposing RB1 mutation should therefore be proposed to all Rb patients.2, 3 The pattern of mutations found in molecular studies revealed the existence of 6% of complete deletions of the RB1 gene that are associated with variable phenotypes.4, 5
Interstitial 13q deletions involving RB1 and its flanking regions, initially revealed by karyotype analyses in Rb patients, were found to be associated with dysmorphic, cranial and hand/foot abnormalities, psychomotor delay and hypotonia.6, 7 Correlations between the size of the deletion and the phenotype were therefore investigated. Although a correlation between the size of the deletion and a specific pattern of malformations and dysmorphism was not established, psychomotor delay was suspected to be restricted to patients harbouring a deletion that encompasses more than the 13q14 band.7 The size and location of the deletion may therefore define the risk of psychomotor delay in a context of contiguous gene syndrome as previously demonstrated, for example, in neurofibromatosis type 1.8 The correlation between the size of the deletion and psychomotor delay in Rb has not yet been determined because of the limited resolution of karyotype analysis. High-resolution analysis of deletions, for example, by CGH, allows this issue to be properly addressed. This work is a nice follow-up of previous studies,9, 10 as it specifically tackles for the first time the issue of psychomotor delay in RB1-deleted patients. We used a dedicated RB1-customized CGH-array designed to define a critical interval and consequently identify candidate genes. This study also provides clues concerning the role of CGH array in Rb molecular diagnosis and parent/patient information regarding genetic counselling.
Patients and methods
Patients
Diagnosis of Rb was established on the basis of examinations by an ophthalmologist and a paediatrician, and by histopathological criteria when the tumour was available. Rb patients were offered genetic counselling, and individual written consent was obtained from all sampled individuals or their legal guardians. In this series of 1160 consecutively ascertained cases, 320 mutations were found, and a total of 17 patients were diagnosed with a complete deletion of the RB1 gene by QMPSF or karyotype analyses. A sufficient amount of DNA was available for CGH analysis in 12 of these patients. Psychomotor delay was reported either when a paediatrician, geneticist and/or psychometrician observed a delayed motor development or speech acquisition delay, or when the patient was taken into care by a specialized educational structure (reported in Table 1 as a binary variable ‘yes’ or ‘no’).
Cytogenetic analysis
Karyotype analyses with RHG banding and FISH with an RB1 probe (Vysis, Downers Grove, IL, USA) were performed according to standard cytogenetic procedures. A customized CGH array centred on the RB1 locus was designed on a 1 × 385-K oligonucleotide CGH microarray (Roche NimbleGen, Madison, WI, USA). The covered region corresponded to the genomic position Chr13:34000000–74000000 (Hg18), for example, a 100-bp resolution. Data were analysed using VAMP software.11
Characterization and sequencing of breakpoints
First, MP/LC12, 13 was used to refine CGH analysis. MP/LC is a technique for the detection of chromosomal rearrangements, which combines the advantages of semiquantitative multiplex PCR and quality of separation of DHPLC. Long-range PCRs were then performed using TripleMaster PCR System (Eppendorf, Hamburg, Germany). Amplicons were sequenced using the BigDye Terminator V1.1 Cycle Sequencing Ready Reaction kit (Life Technologies, Carlsbad, CA, USA), followed by electrophoresis in an ABI 3130xl (Life Technologies).
Results
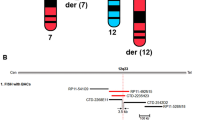
The 13q deletion in all 12 patients was characterized by our RB1-customized CGH array (Table 1 and Figure 1). The largest deletion that was not detected by karyotype analysis measured 8.2 Mb and the smallest deletion detected by karyotype analysis measured 11 Mb. The karyotype resolution was therefore about 10 Mb, which is consistent with routine diagnostic practice. The sequencing experiment (Figure 2) demonstrated good accuracy of the CGH array, as the mean difference of location between the sequencing and CGH mapping results was equal to 1.08 kb (±1.7 kb, SD). Unfortunately, long-range PCRs and breakpoint sequencing failed in five cases due to low complexity and/or repeated regions. Nevertheless, CGH resolution was sufficient to allow breakpoint location, for example, for PCDH9 (see below).
CGH array results from 12 Rb patients harbouring a complete RB1 deletion. Adapted view from UCSC. From top to bottom, schematic view of chromosome, deletion mapped by CGH array labelled with the patient number, representation of cytogenetic bands and Refseq genes. Deletions represented by a grey bar were excluded from the definition of ‘the zone of interest’ (flanked by the black frame) because of mosaicism or confounding diagnosis. PCDH8 loss was always associated with psychomotor delay, whereas a deletion encompassing NUFIP1 cosegregated in patient 6 and his affected relatives, without any detectable impact on psychomotor development (see text for details). The Refseq genes shown are only those cited in the text, as well as all those included in the zone of interest.
Breakpoint sequencing results. Sequencing electrophoregrams of the deletion breakpoints found in seven patients. The upper part represents an adapted UCSC view in which a schematic view of the chromosome and a Refseq gene representation are included, corresponding to the sequencing electrophoregram. The vertical black line represents the breakpoints.
Patients 1 to 6 presented a molecular microdeletion (not detected by karyotype analysis), and patients 7 to 12 presented a cytogenetic deletion centred around the 13q14.2 band. One of the 6 patients with a molecular microdeletion and 5 of the 6 patients with a cytogenetic deletion presented psychomotor delay (Table 1). Cytogenetic deletions in a context of Rb were therefore associated with psychomotor delay (P=0.03; Fisher’s exact test; two-sided). To define the minimal deletion associated with psychomotor delay, patient 5 (with psychomotor delay) and patient 8 (without psychomotor delay) were excluded from the following analysis because they presented documented fetal suffering or mosaicism documented by karyotype, respectively (Table 1; see Discussion).
The largest deletion found without psychomotor delay measured 8.2 Mb (Table 1, patient 6). Sequencing analysis (Figure 2) showed that the breakpoint were located inside the EPSTI1 and FAM124 genes. This interval (chr13:42449743_50715540) of deletion was inherited from the patient’s father. In patients 2 and 4, who did not present psychomotor delay, sequencing analysis localized breakpoints inside the DLEU2 and CDACC1 genes. The deletion identified in patient 7 (chr13:13213373_54501464) was the smallest deletion (11.2 Mb) associated with psychomotor delay. The breakpoint was found inside the ENOX1 gene (Figure 2). Combining these results with those from patient 6, the chr13:50715540_54501464 interval defined an RB1-flanking telomeric region where candidate genes for psychomotor delay may be found (Figure 1). Twenty-five Refseq genes have been reported inside this region, including PCDH8 (Supplementary Table 1).
Discussion
High-resolution CGH reliably defined a deleted interval not associated with psychomotor delay (chr13:42449743_50715540; Table 1 and Figure 1). This deletion was inherited (Table 1), conferring a high degree of confidence to our results, by excluding the mosaicism bias. On the other hand, mosaicism led to exclusion of patient 8, as a mosaic status can be associated with an attenuated phenotype and should not be considered in terms of the genotype/phenotype relationship.14 As Rb is a disease with a high rate of de novo mutations, an attenuated phenotype (ie, an absence of psychomotor delay in a patient harbouring a cytogenetic deletion) in first-generation mutation carriers is not unexpected.15, 16 Patient 5 was also excluded from the analysis because fetal suffering can be responsible for psychomotor delay, thereby introducing another analysis bias.
The breakpoints of the deletion located in patients with normal development demonstrated that DLEU2, CDACC1, EPST1 and FAM124A genes, disrupted by the deletion, therefore cannot be associated with psychomotor delay. A breakpoint in the ENOX1 gene was also found in a patient with psychomotor delay. Inhibition of ENOX1 has been reported to decrease angiogenesis in tumour growth.17 ENOX1 was found with a high but not exclusive expression level in fetal brain (see http://biogps.org), but no other data are available in the literature to incriminate this gene in the context of psychomotor delay. Comparative analysis of deleted intervals in delayed and non-delayed patients identified PCDH8 as a candidate gene for psychomotor delay (Figure 1). PCDH8 (MIM no. 603580), for protocadherin 8, is located in 13q14.3, and belongs to a subclass of cadherins.18 PCDH8 has a brain-specific expression making this gene a good candidate gene for psychomotor delay. Furthermore, previous linkage data suggested PCDH8 as a candidate gene for schizophrenia.19 Also of interest is that protocadherin PCDH19 has been previously involved in the female-restricted epilepsy and mental retardation syndrome.20 Interestingly, 2 patients (Figure 1; patient 7 and 11) with psychomotor impairment and loss of a PCDH8 copy suffered from epilepsy. Also, our data clearly point to PCDH8 as one of the putative genes responsible for psychomotor delay in the context of Rb, acting either directly or indirectly via regulatory mechanisms. An autosomic dominant model linked to PCDH8 loss-of-function could be suspected, but a recessive model driven by epigenetic inactivation of the second allele of PCDH8 cannot be excluded, as the PCDH8 promoter has been found to be methylated.21 Two previous studies also suggested the role of PCDH8, but did not formally map this gene, as confounding factors such as mosaicism and alternative causes of psychomotor delay were not evaluated.9, 10 Nevertheless, one study described a few deleted patients without PCDH8 involvement, who did present psychomotor delay, leading authors to designate NUFIP1 as another possible candidate gene.10 Overall data show that loss of neither PCDH8 nor NUFIP1 can explain all delayed cases, but PCDH8 loss is always associated with psychomotor delay, as opposed to NUFIP1 (see patient 6, Figure 1).
Another interesting finding was that the deletion breakpoints in patient 9, who presented psychomotor delay, were located inside the PCDH9 gene (Figure 1). On the basis of the role of protocadherins in neuronal development and neuronal plasticity,22 a PCDH dose/effect in the expression of psychomotor delay, implying genetic heterogeneity, could be proposed in the context of complete RB1 deletion.
In summary, we demonstrated that loss of PCDH8 in the context of complete deletion of RB1 should alert geneticists to the risk of psychomotor delay. Fine mapping of deletion breakpoints is therefore mandatory in Rb patients in case of the following: (i) complete or partial RB1 deletion encompassing a flanking region and (ii) mental delay either isolated or associated with dysmorphic features. This could be performed by CGH array of the chromosome 13 or, in the near future, by a global approach such as massively parallel sequencing. This second line of investigation will precisely define the deleted genes flanking RB1, and thereby improve genetic counselling/information and define the most appropriate follow-up options.
References
Friend SH, Bernards R, Rogelj S et al. A human DNA segment with properties of the gene that predisposes to retinoblastoma and osteosarcoma. Nature 1986; 323: 643–646.
Knudson AG : Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci USA 1971; 68: 820–823.
Comings DE : A general theory of carcinogenesis. Proc Natl Acad Sci USA 1973; 70: 3324–3328.
Albrecht P, Ansperger-Rescher B, Schuler A, Zeschnigk M, Gallie B, Lohmann DR : Spectrum of gross deletions and insertions in the RB1 gene in patients with retinoblastoma and association with phenotypic expression. Hum Mutat 2005; 26: 437–445.
Houdayer C, Gauthier-Villars M, Lauge A et al. Comprehensive screening for constitutional RB1 mutations by DHPLC and QMPSF. Hum Mutat 2004; 23: 193–202.
Bunin GR, Emanuel BS, Meadows AT, Buckley JD, Woods WG, Hammond GD : Frequency of 13q abnormalities among 203 patients with retinoblastoma. J Natl Cancer Inst 1989; 81: 370–374.
Baud O, Cormier-Daire V, Lyonnet S, Desjardins L, Turleau C, Doz F : Dysmorphic phenotype and neurological impairment in 22 retinoblastoma patients with constitutional cytogenetic 13q deletion. Clin Genet 1999; 55: 478–482.
Pasmant E, Sabbagh A, Spurlock G et al. NF1 microdeletions in neurofibromatosis type 1: from genotype to phenotype. Hum Mutat 31: E1506–E1518.
Caselli R, Speciale C, Pescucci C et al. Retinoblastoma and mental retardation microdeletion syndrome: clinical characterization and molecular dissection using array CGH. J Hum Genet 2007; 52: 535–542.
Mitter D, Ullmann R, Muradyan A et al. Genotype-phenotype correlations in patients with retinoblastoma and interstitial 13q deletions. Eur J Hum Genet 2011; 19: 947–958.
La Rosa P, Viara E, Hupe P et al. VAMP: visualization and analysis of array-CGH, transcriptome and other molecular profiles. Bioinformatics 2006; 22: 2066–2073.
Dehainault C, Lauge A, Caux-Moncoutier V et al. Multiplex PCR/liquid chromatography assay for detection of gene rearrangements: application to RB1 gene. Nucleic Acids Res 2004; 32: e139.
Delnatte C, Sanlaville D, Mougenot JF et al. Contiguous gene deletion within chromosome arm 10q is associated with juvenile polyposis of infancy, reflecting cooperation between the BMPR1A and PTEN tumor-suppressor genes. Am J Hum Genet 2006; 78: 1066–1074.
Taylor M, Dehainault C, Desjardins L et al. Genotype-phenotype correlations in hereditary familial retinoblastoma. Hum Mutat 2007; 28: 284–293.
Sippel KC, Fraioli RE, Smith GD et al. Frequency of somatic and germ-line mosaicism in retinoblastoma: implications for genetic counseling. Am J Hum Genet 1998; 62: 610–619.
Castera L, Sabbagh A, Dehainault C et al. MDM2 as a modifier gene in retinoblastoma. J Natl Cancer Inst 2010; 102: 1805–1808.
Geng L, Rachakonda G, Morre DJ et al. Indolyl-quinuclidinols inhibit ENOX activity and endothelial cell morphogenesis while enhancing radiation-mediated control of tumor vasculature. FASEB J 2009; 23: 2986–2995.
Strehl S, Glatt K, Liu QM, Glatt H, Lalande M : Characterization of two novel protocadherins (PCDH8 and PCDH9) localized on human chromosome 13 and mouse chromosome 14. Genomics 1998; 53: 81–89.
Bray NJ, Kirov G, Owen RJ et al. Screening the human protocadherin 8 (PCDH8) gene in schizophrenia. Genes Brain Behav 2002; 1: 187–191.
Jamal SM, Basran RK, Newton S, Wang Z, Milunsky JM : Novel de novo PCDH19 mutations in three unrelated females with epilepsy female restricted mental retardation syndrome. Am J Med Genet A 2010; 152A: 2475–2481.
Yu JS, Koujak S, Nagase S et al. PCDH8, the human homolog of PAPC, is a candidate tumor suppressor of breast cancer. Oncogene 2008; 27: 4657–4665.
Kim SY, Mo JW, Han S et al. The expression of non-clustered protocadherins in adult rat hippocampal formation and the connecting brain regions. Neuroscience 2010; 170: 189–199.
Acknowledgements
This work was supported by grants from the ‘Programme Incitatif et Coopératif sur le Rétinoblastome’ (Institut Curie) and RETINOSTOP. We thank Laurence Desjardins, Virginie Moncoutier, Carole Tirapo, Catherine Dubois d’Enghien, Anthony Laugé, Isabelle Eugène, Sandrine Miglierina, Catherine Rougeron and Catherine Gilbon for their helpful support during the study. We also thank clinicians from the GGC (Groupe Génétique et Cancer) for referring patients.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Castéra, L., Dehainault, C., Michaux, D. et al. Fine mapping of whole RB1 gene deletions in retinoblastoma patients confirms PCDH8 as a candidate gene for psychomotor delay. Eur J Hum Genet 21, 460–464 (2013). https://doi.org/10.1038/ejhg.2012.186
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2012.186
Keywords
This article is cited by
-
Genetic Testing for Eye Diseases: A Comprehensive Guide and Review of Ocular Genetic Manifestations from Anterior Segment Malformation to Retinal Dystrophy
Current Genetic Medicine Reports (2016)