Abstract
Hydatidiform mole (HM) is an abnormal human pregnancy, where the placenta presents with vesicular swelling of the chorionic villi. A fetus is either not present, or malformed and not viable. Most moles are diploid androgenetic as if one spermatozoon fertilized an empty oocyte, or triploid with one maternal and two paternal chromosome sets as if two spermatozoa fertilized a normal oocyte. However, diploid moles with both paternal and maternal markers of the nuclear genome have been reported. Among 162 consecutively collected diploid moles, we have earlier found indications of both maternal and paternal genomes in 11. In the present study, we have performed detailed analysis of DNA-markers in tissue and single cells from these 11 HMs. In 3/11, we identified one biparental cell population only, whereas in 8/11, we demonstrated mosaicism: one biparental cell population and one androgenetic cell population. One mosaic mole was followed by persistent trophoblastic disease (PTD). In seven of the mosaics, one spermatozoon appeared to have contributed to the genomes of both cell types. Our observations make it likely that mosaic conceptuses, encompassing an androgenetic cell population, result from various postzygotic abnormalities, including paternal pronuclear duplication, asymmetric cytokinesis, and postzygotic diploidization. This corroborates the suggestion that fertilization of an empty egg is not mandatory for the creation of an androgenetic cell population. Future studies of mosaic conceptuses may disclose details about fertilization, early cell divisions and differentiation. Apparently, only a minority of diploid moles with both paternal and maternal markers are ‘genuine’ diploid biparental moles (DiBiparHMs).
Similar content being viewed by others
Introduction
Hydatidiform mole (HM) is an abnormal human pregnancy, characterized by vesicular swelling of the chorionic villi and hyperplasia of the trophoblastic layer. By far, most HMs are diploid or triploid. In most diploid HMs, analyses of markers of the nuclear genome identify markers that are identical with markers in the paternal genome only (diploid androgenetic HMs, DiAndHMs). Most DiAndHMs show homozygosity in all loci analyzed, as if an empty oocyte was fertilized by one spermatozoon, followed by duplication of the haploid paternal genome. Fewer DiAndHMs show heterozygosity in some loci as if an empty oocyte was fertilized by two independent spermatozoa from the same father.1 However, over the years, diploid HMs with biparental contributions to the genome have been reported.2, 3, 4, 5, 6, 7
A molar phenotype in a diploid conceptus with biparental alleles may be caused by paternal methylation patterns on maternally inherited chromosomes.8, 9, 10, 11 Another possible explanation is the co-existence of two diploid cell populations, one androgenetic and the other biparental.12, 13, 14, 15, 16
In The Danish Mole Project, we have consecutively been collecting samples from HMs since 1986. In the present study, we have subjected 11 diploid HMs showing signs of having biparental genomes to detailed genetic analyses, and found indications that only a minority of these have ‘true’ biparental genomes, whereas most are mosaics.
Materials and methods
In the Danish Mole Project, fresh tissue is collected from conceptuses presenting with vesicular villi. In the period April 1986–June 2003, samples from 309 conceptuses were received. Previously, material was successfully retrieved for histopathological revision from 294 of these conceptuses, and 270 samples were classified as originating in HMs. Ploidy was determined by karyotyping of uncultured and/or cultured cells, and/or by measurement of the nuclear DNA contents by flow cytometry of unfixed nuclei. The parental origin of the genome was determined by comparing DNA markers in the moles with those in the parents. Of the 270 HMs, 162 were diploid, and of these, 11 were classified as having genomes from both parents.17
In the present work, DNA from tissue samples from the 11 moles, showing signs of having genome from both parent, and DNA from the parents were subjected to detailed analysis, using a panel of at least 10 microsatellite markers. The patterns of peaks in the molar electropherograms were interpreted by comparing with the electropherograms that would be expected for various hypothetical offspring of the parents: a diploid androgenetic conceptus, a diploid biparental conceptus, and a mixture of these two. Rough estimates of the frequencies of the androgenetic cells were made by visual evaluation of the heights of the peaks.18
From four moles, DNA from single cells was prepared by incubating unfixed vesicular villi with collagenase D. After rinsing five times in cell culture medium, DNA was prepared by incubation with proteinase K, and amplified with primers for the markers D6S105 and D6S2443, in a multiplexed analysis.
For details on materials, methods, and interpretation, see Supplementary Materials and Methods, and Supplementary Table 1, online).
Clinical data were obtained from a questionnaire filled in by the parents and from the medical records.
The regional Committee on Biomedical Research Ethics approved the study. All participants gave informed consent.
Results
Eleven diploid HMs displaying both maternal and paternal alleles, identified in a consecutive cohort of 270 HMs, were subjected to genetic analyses. The results are summarized in Table 1, along with clinical and morphological data. The mother of HM131 has had a total of six pregnancies, all showing molar morphology. She is a member of a consanguineous family, and she is the first cousin of two sisters with repetitive diploid moles with alleles from both parents. Out of the molar pregnancies of this patient and her relatives, only HM131 was part of the cohort of 270 HMs. Observations in this family have been published before.6, 19, 20 The parents of the 10 other moles were Danish, and had no personal or familial history of moles.
In four HMs, both uncultured and cultured cells were karyotyped. In two of these, we observed a discrepancy between the karyotypes of uncultured cells and cultured cells. In HM269, the karyotype in uncultured cells was 46,XY, whereas the karyotype after culture was 46,XX. In HM192 the uncultured cells had a surplus derivative chromosome 7, which was not found in the cultured cells. In both HMs, comparison of chromosomal heteromorphisms made it unlikely that these discrepancies were caused by maternal overgrowth in the cell culture.
Analyzing microsatellite markers we found imbalances in the signals in eight of the 11 HMs. None of the moles had more than one maternally derived allele at any locus, excluding maternal contamination. Rather, we either observed a disproportion between the heights of the peaks representing two alleles (Figures 1a–c) and/or three peaks at the same locus (Figures 1b and c). See Supplementary Table 1, online, for details. In all cases, the third peak, or the surplus height of one or two peak(s), represented one or both paternal allele(s). The most probable explanation of these findings is the presence of two cell populations, one androgenetic cell population and one cell population with balanced biparental contributions to the genome (DiAnd/BiparHM). Among the eight DiAnd/BiparHMs, the androgenetic cell population appeared homozygous in four moles. In three of these, the paternal allele found in the androgenetic cell population was identical with the paternal allele in the biparental cell population in all loci analyzed (P1M+P1P1), whereas in one case, the paternal allele found in the androgenetic cell population was different from the paternal allele in the biparental cell population in several loci (P1M+P2P2). In four cases, the androgenetic cell population was heterozygous (P1M+P1P2) (Table 1).
Microsatellite markers in hydatidiform moles (HMs) showing both paternal and maternal markers. Pat., alleles in the father; Mat., alleles in the mother; HM-1, alleles in the molar tissue, first sample; HM-2, alleles in the molar tissue, second sample; green peaks, D13S258; blue peaks, D18S51. Broad blue peaks at 240 bp in some analyses are artefacts. Within some peaks, smaller artefact peaks of a different color are seen. (a) HM142 (P1M+P1P1); (b) HM269 (P1M+P2P2); (c) HM192 (P1M+P1P2); (d) HM635 (PM).
Because the observation of two cell populations could be caused by analyzing a mixture of tissues from both conceptuses of a twin pregnancy, we repeated the microsatellite analysis on DNA prepared from a second specimen, paying special attention to using only one, integral, sample of vesicular villi. In seven of the eight HMs classified as DiAnd/BiparHMs in the first experiment, the same two cell populations were identified in the second analysis, whereas in the second sample from HM192, only the androgenetic cell population was identified (Figure 1 and Supplementary Table 1, online). Rough estimates of the frequencies of the two types of cells are given in Figure 2.
Relative frequencies of the androgenetic cells in the first and second sample from 11 diploid hydatidiform moles (HMs) with paternal and maternal contribution to the genome. The first sample of DNA was prepared from representative parts of the chorionic villi. The second sample of DNA was prepared from one integral piece of vesicular villi. The relative frequencies of the androgenetic cells in the two different tissue samples were estimated by visual inspection of the peaks for all informative loci, and categorized as: 0%, no androgenetic cells; 25%, androgenetic cells present, but less frequent than the biparental cells; 50%, androgenetic cells and biparental cells equally frequent; 75%, androgenetic cells more frequent than biparental cells; 100%, exclusively androgenetic cells.
To further substantiate the existence of two cell populations, we co-analyzed the microsatellite markers D6S105 and D6S2443 in single cells from four DiAnd/BiparHMs (Table 2, Figure 3, and Supplementary Table 2, online). For three HMs, a total of 33/43 single cells gave unequivocal results: 31 cells (94%) were biparental and two cells (6%) were androgenetic. For HM173, none of the 25 cells gave unequivocal results. We therefore supplemented with PCR, using the fluid around the single cells of HM173 as template. In all of the five samples from the fluid of the unrinsed suspension of single cells, we saw the same alleles as in DNA isolated from molar tissue, whereas PCR on 18 samples of the fluid from the last rinse procedure gave rise to an unsystematic combination of alleles present and alleles not present in the molar tissue.
Microsatellite markers in single cells from a hydatidiform mole (HM) showing both paternal and maternal markers: HM539 (P1M + P1P2). DNA from the father (Pat), mother (Mat), and from tissue and two different single cells from the mole were co-analyzed in the loci D6S105 and D6S2443. The alleles in polymorphic locus D6S105 are represented by fragments in the range 117–121 bp. The alleles in the polymorphic locus D6S2443 are represented by fragments in the range 165–193 bp. In the molar tissue, both a biparental cell population, and an androgenetic cell population can be identified. The two single cells represent the biparental and the androgenetic cell population, respectively. (Due to overload of the PRC product in the analysis of DNA from the father and the molar tissue, the peaks for the 121 allele at D6S105, and for the 165 and 175 alleles at D6S2443 are truncated.)
In three moles (HM101, HM131, and HM635), analyses of DNA from both tissue samples identified one biparental cell population, only. (Figures 1d and 2, Table 1). These moles may be ‘genuine’ diploid biparental moles (DiBiparHMs).
The histopathological diagnoses were revised as part of a previous study.17 In all the 11 moles analyzed in the present study, trophoblastic hyperplasia was confirmed. No sign of fetal differentiation was observed in any of the 11 moles by ultrasound, at the histopathological examination, or at the inspection of the tissue received for genetic analyses. Neither was any sign of placental mesenchymal dysplasia (PMD) reported. Out of our original cohort of 162 consecutive diploid HMs, eight were part of a multiple pregnancy. None of the 11 diploid moles with markers from both parents was among these.
The morphologic diagnoses made by the local pathologists and by the reviewing pathologist are listed in Table 1. Both pathologists classified a substantial fraction of these 11 diploid moles as PHMs, and this diagnosis was used both in the group of DiAnd/BiparHMs, and in the group of DiBiparHMs. In four cases, the two pathologists made different diagnoses.
Discussion
In eight of 11 HMs with biparental genomic markers (73%), we found indications of two cell populations, one androgenetic and one biparental (DiAnd/BiparHMs), whereas three (27%) appeared to be ‘genuine’ DiBiparHMs. The study is the most representative hereto, as it is based on the largest cohort of consecutively collected, fresh samples from HMs. As exclusively morphologic criteria are used for including samples in the Danish Mole Project, we could evaluate the genetic causes of the molar phenotype in an unbiased way.
Two cell populations
One possible explanation for the observation of markers from two cell populations is that tissues from both conceptuses of an unrecognized twin pregnancy were mixed at evacuation. However, in the present cases, this is unlikely. In none of the diploid HMs with biparental genomic markers, fetal differentiation was noted. Moreover, when we repeated DNA preparation, paying special attention to exclusively using one integral sample of vesicular villi, the same two cell populations were identified in seven of the eight HMs.
We attempted to further document the existence of two cell populations by analyzing single cells. The observation of a number of different allele patterns made the interpretation of these analyses challenging. The observation of alleles not present in the molar tissue indicates that, when we intended to analyze samples with a total of two template molecules (ie, the genome of one single cell), or no template molecule (the rinse fluid), DNA from various sources could serve as template in the PCR reaction. Thus, we cannot exclude that in some cases, allele patterns observed in single cells that were identical with allele patterns in one of the predicted cell populations, actually were caused by contamination. In addition, the observation of only one allele in the PRC product could be caused by selective amplification of one allele of a heterozygous cell (allele dropout).
Despite these limitations, we found clear evidence for the biparental cell population predicted by the analysis of DNA from villous tissue in three HMs. In contrast, significantly fewer cells than expected showed the allele pattern of the androgenetic cell population. As a limited number of single cells were isolated, this could be caused by chance. Alternatively, the androgenetic nuclei may have been most frequent in the syncytiotrophoblastic cells, which would not be represented in a single cell suspension.
The observation of a large variety of allele patterns in single cells from HM173 is at present unexplained. Although the results of analysis of DNA from villous tissue could be explained by the existence of two diploid cell populations, it cannot be excluded that HM173 actually contained more than two cell populations. Another possible explanation is that the tissue of HM173 was more degraded than the other three HMs analyzed, leading to a more unfavorable balance between analyzable DNA from the single cells and free DNA from various sources.
Diploid moles may originate in triploid zygotes
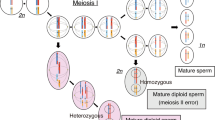
Golubovsky21 highlighted that fertilization of one oocyte by two spermatozoa, followed by abnormal division of the zygote and endoreduplication of one pronucleus (post-zygotic diploidisation) could give rise to various combinations of cell populations. In five of the eight DiAnd/BiparHMs, we found alleles consistent with diploidization of a triploid zygote, arisen by dispermy: In one of these cases, two duplicated (identical) paternal genome sets could have made possible the androgenetic cell population (Figure 4a), whereas in four cases, the nuclear genome in the androgenetic cell population seems to have originated in two different paternal pronuclei (Figure 4b), or by fertilization by three spermatozoa (not illustrated). Three cases could have arisen by fertilization of one oocyte by one spermatozoon, followed by duplication of the paternal pronucleus, creating a ‘temporary tripronuclear zygote’ with two identical paternal pronuclei, before post-zygotic diploidization (Figure 4c).
Possible fertilizations, endoreduplications, and abnormal cell divisions in mosaic hydatidiform moles (HMs), and twin gestations including an HM. (a) Fertilisation by two sperms; one giving rise to the paternal genome set in the diploid biparental cell population, the other giving rise to both genome sets in the diploid androgenetic cell population via endoreduplication. (b) Fertilisation by two sperms; one giving rise to one of the paternal genome sets in the diploid androgenetic cell population, the other contributing one genome set to both cell populations via endoreduplication. (c) Fertilisation by one sperm that via two endoreduplications gives rise to three identical paternal genome sets, of which two constitute the genome of the diploid androgenetic cell population and one is the paternal genome set in the diploid biparental cell population.
We previously analyzed seven twin pregnancies consisting of a diploid HM and a normal biparental placenta with a fetus, and found indications of the same mechanisms.22 However, whereas in the twin gestations the nuclear genome in the androgenetic molar placenta most frequently was homozygous and different from the paternal genome set in the biparental conceptus (Figure 4a), in most of the mosaic cases, the paternal genome set in the biparental cells was also present in the androgenetic cells (Figures 4b and c). As numbers are small, this may have occurred by chance. However, identical paternal genome sets can only be present in both cell populations if at least one abnormal duplication of a paternal genome set took place before the zygote underwent the first cell division (Figures 4b and c), whereas in the case of two cell populations with different paternal genome sets, the duplication may have taken place after the first cell division (Figure 4a). A delayed first cell division may increase the probability of endoreduplication prior to cell division, and thus the ability of both cell populations to proliferate and stay together as one entity.
As suggested by others, a biparental cell population may be present, along with the androgenetic cell population in moles classified as DiAndHMs.12, 21 This prediction is verified by the fact that we would have overlooked the biparental cell population in one case (HM192), had we analyzed the second sample of vesicular villi, only.
Mosaicism: HM versus PMD versus fetal malformation
In the DiAnd/BiparHMs, the molar phenotype may be ascribed to the differentiation of the androgenetic cells. Indications of mosaicism/chimerism with an androgenetic and a biparental cell population have been observed both in HMs,12, 13, 14, 15, 16, present study in placentas displaying PMD,18, 23, 24, 25 and in fetuses/children with malformations or growth abnormalities mimicking (part of) the Beckwith–Wiedemann phenotype.25, 26, 27, 28, 29, 30 The phenotype seems to correlate with the localization of the androgenetic cells. In the cases presenting with malformations in a fetus/child, the androgenetic cells were observed in the fetus/child, and in PMD, the androgenetic cells have been observed predominantly in the placental vessels, chorion, and mesenchymal cells. In an HM with two cell populations, androgenetic cells presented after direct harvest, but not after subcultivation, suggesting that these cells were present in the trophoblastic layer, but not in the mesenchymal cells.12 Accordingly, we observed a low frequency of androgenetic cells among single cells from mosaic HMs, suggesting that these cells were most frequent in the syncytiotrophoblastic layer. However, like others,14 we identified androgenetic markers, only, in DNA isolated from some chorionic villi of a conceptus with both an androgenetic and a biparental cell population, indicating that the androgenetic cells were present both in the trophoblastic layer and in the stroma. In addition, androgenetic cells have been found in amniotic tissue of molar conceptuses.13 Possibly, the crucial factor for what presents as an HM, and what presents as a PMD, is not the absolute presence/absence of androgenetic cells within certain parts of the placenta, but rather the relative frequencies of androgenetic and biparental cells. Accordingly, we should expect to see conceptuses that show various mixtures of the mesenchymal dysplasia phenotype, the molar phenotype, and/or a fetus with developmental disturbances. Indeed, a mosaic conceptus presenting as a partial mole with a hermaphroditic fetus has been reported.15 In The Danish Mole Project, we ascertain for placentas displaying molar morphology. That may explain why neither fetal differentiation nor PMD were observed in any of our eight DiAnd/BiparHMs.
Mosaicism versus genuine diploid biparental conceptuses
In three of the 11 diploid HMs with biparental markers, we identified a biparental cell population, only (DiBiparHMs): HM101, HM131, and HM635. One could speculate if all diploid HMs with biparental markers actually are DiAnd/BiparHMs, and that in HMs presenting as DiBiparHMs the androgenetic cell population was overlooked, for instance by selection of the tissue for marker analysis. In a pregnancy with an unremarkable fetus and intermixed populations of morphologically normal chorionic villi and villi with molar morphology, an androgenetic cell population would have been overlooked if only normally appearing villi had been analyzed.14 However, in an abnormal conceptus, it is unlikely that the abnormal tissue would not be examined. Furthermore, in our three moles with balanced biparental alleles, we also observed the balanced biparental alleles when we repeated the analysis in DNA from vesicular villi, only. We thus regard it highly unlikely that the molar morphology of these conceptuses was caused by a co-existing androgenetic cell population. Rather, these three conceptuses were ‘genuine’DiBiparHMs.
We thus find it most likely that ‘diploid HMs with biparental markers’ consists of two different entities: (1) conceptuses with two cell populations (DiAnd/BiparHMs), the phenotype caused by the androgenetic cell population and (2) DiBiparHMs, the molar phenotype caused by other factors such as abnormal parental imprinting.
In parallel, some cases of PMD may be caused by the co-existence of an androgenetic cell population and a biparental cell population, whereas other cases may be caused by other factors. In a number of cases, such two cell populations have been demonstrated,18, 23, 24, 31 whereas we previously found identical biparental contributions to the genome of a placenta with PMD, and to the genome of the healthy fetus.32
Persistent trophoblastic disease (PTD)
The most important clinical issue is the risk of persistent trophoblastic disease (PTD). PTD can follow both a molar and a non-molar pregnancy. The average risk of PTD after an HM is 10%, the risk almost exclusively adhering to the diploid moles.17 The risk of PTD after a non-molar pregnancy is low.33 It has been assumed that half the cases of choriocarcinoma succeed an HM, and half succeed a non-molar conception.34 However, in a recent study of eight gestational choriocarcinomas, androgenetic origin of the malignant cells was demonstrated in six cases, although only one of these women had an obvious molar pregnancy history.35 We observed PTD after one of the eight DiAnd/BiparHMs, and after none of the three DiBiparHMs. PTD has also been observed after a case of PMD with an androgenetic and a biparental cell population,23 and after a case of hermaphroditism and partial mole.15 Furthermore, an androgenetic cell line was identified in a Wilm's tumor, and in other tissues of a patient with Beckwith–Wiedemann syndrome.30 Thus, the malignant potential may not be related to the molar morphology, per se, but rather to the presence of an androgenetic cell population. The frequency of an androgenetic cell population in non-molar conceptuses is unknown, but probably low. However, androgenetic cells have been observed so frequently in PMD that it seems wise to recommend measurements of se-hCG after this rare condition. Evaluation of the risk in pregnancies where the fetus is malformed or has growth abnormalities is more complicated, as there may be many different causes. However, as androgenetic cells have been observed in fetuses/persons with hermaphroditism, and in fetuses/persons with a Beckwith–Wiedemann-like phenotype, it seems reasonable to offer hCG surveillance to women who have had such pregnancies, along with genetic investigation and counseling to the woman and her children.
Morphology
Although the main aim of the present study was to explore the genetic constitution of molar conceptuses with both maternal and paternal markers, we have also listed the morphologic diagnoses made by the local pathologists and the revising pathologist. The two pathologists made their diagnoses under different conditions. The local pathologist could inspect the evacuated tissue macroscopically and choose which parts to inspect microscopically, whereas the revising pathologist made her diagnosis from the blocks/sections forwarded for revision. Also, as we included only conceptuses diagnosed as HM by the revising pathologist, our cohort may be biased against conceptuses where the local pathologist noted a more ‘molar phenotype,’ than the revising pathologist. Despite these limitations, the high frequency of disagreement in the morphologic diagnoses is remarkable. In both genetic subgroups, both pathologists classified some moles as PHMs and some as CHMs, and in four cases, the two pathologists made different diagnoses. In two cases the revising pathologist noted a less ‘molar phenotype’ than the local pathologist (PHM versus CHM), and in two cases, the revising pathologist noted a more ‘molar phenotype’ (PHM versus hydropic abortion and ‘hydatidiform mole?’, respectively). Usually, a substantial correlation between phenotype and genotype is reported (CHMs mostly being diploid, and PHMs mostly being triploid).1 However, possibly the criteria for the morphologic subclassification of HMs are not optimal for the rare DiAnd/BiparHMs and DiBiparHMs. Also, at least for the mosaics, the selection of tissue for examination may influence the diagnosis made.
Conclusion
In a significant fraction of diploid HMs showing genetic markers from both parents, the mole consists of two cell populations, one androgenetic and one biparental (DiAnd/BiparHMs). It is possible that the mosaicism arise by abnormal duplication of chromosomes and/or abnormal cell division in tripronuclear conceptuses. Genuine DiBiparHMs seem to be less frequent. In future studies of diploid moles with biparental markers, one should discriminate between these two types. Further studies of DiAnd/BiparHMs could disclose details about fertilization, early cell divisions, and differentiation. Studies of DiBiparHMs may disclose human genes regulated by genomic imprinting.
References
Slim R, Mehio A : The genetics of hydatidiform moles: new lights on an ancient disease. Clin Genet 2007; 71: 25–34.
Jacobs PA, Wilson CM, Sprenkle JA et al: Mechanism of origin of complete hydatidiform moles. Nature 1980; 286: 714–716.
Vejerslev LO, Fisher RA, Surti U et al: Hydatidiform mole: cytogenetically unusual cases and their implications for the present classification. Am J Obstet Gynecol 1987; 157: 180–184.
Ko TM, Hsieh CY, Ho HN et al: Restriction fragment length polymorphism analysis to study the genetic origin of complete hydatidiform mole. Am J Obstet Gynecol 1991; 164: 901–906.
Kovacs BW, Shahbahrami B, Tast DE et al: Molecular genetic analysis of complete hydatidiform moles. Cancer Genet Cytogenet 1991; 54: 143–152.
Sunde L, Vejerslev LO, Jensen MP et al: Genetic analysis of repeated, biparental, diploid, hydatidiform moles. Cancer Genet Cytogenet 1993; 66: 16–22.
Fisher RA, Paradinas FJ, Soteriou BA et al: Diploid hydatidiform moles with fetal red blood cells in molar villi. 2--Genetics. J Pathol 1997; 181: 189–195.
Judson H, Hayward BE, Sheridan E et al: A global disorder of imprinting in the human female germ line. Nature 2002; 416: 539–542.
El-Maarri O, Seoud M, Coullin P et al: Maternal alleles acquiring paternal methylation patterns in biparental complete hydatidiform moles. Hum Mol Genet 2003; 12: 1405–1413.
Kou YC, Shao L, Peng HH et al: A recurrent intragenic genomic duplication, other novel mutations in NLRP7 and imprinting defects in recurrent biparental hydatidiform moles. Mol Hum Reprod 2008; 14: 33–40.
Hayward BE, De Vos M, Talati N et al: Genetic and epigenetic analysis of recurrent hydatidiform mole. Hum Mutat 2009; 30: E629–E639.
Ford JH, Brown JK, Lew WY et al: Diploid complete hydatidiform mole, mosaic for normally fertilized cells and androgenetic homozygous cells. Case report. Br J Obstet Gynaecol 1986; 93: 1181–1186.
Weaver DT, Fisher RA, Newlands ES et al: Amniotic tissue in complete hydatidiform moles can be androgenetic. J Pathol 2000; 191: 67–70.
Makrydimas G, Sebire NJ, Thornton SE et al: Complete hydatidiform mole and normal live birth: a novel case of confined placental mosaicism: case report. Hum Reprod 2002; 17: 2459–2463.
Shiina H, Oka K, Okane M et al: Coexisting true hermaphroditism and partial hydatidiform mole developing metastatic gestational trophoblastic tumors. A case report. Virchows Arch 2002; 441: 514–518.
Hoffner L, Dunn J, Esposito N et al: P57KIP2 immunostaining and molecular cytogenetics: combined approach aids in diagnosis of morphologically challenging cases with molar phenotype and in detecting androgenetic cell lines in mosaic/chimeric conceptions. Hum Pathol 2008; 39: 63–72.
Niemann I, Hansen ES, Sunde L : The risk of persistent trophoblastic disease after hydatidiform mole classified by morphology and ploidy. Gynecol Oncol 2007; 104: 411–415.
Kaiser-Rogers KA, McFadden DE, Livasy CA et al: Androgenetic/biparental mosaicism causes placental mesenchymal dysplasia. J Med Genet 2006; 43: 187–192.
Helwani MN, Seoud M, Zahed L et al: A familial case of recurrent hydatidiform molar pregnancies with biparental genomic contribution. Hum Genet 1999; 105: 112–115.
Vejerslev LO, Sunde L, Hansen BF et al: Hydatidiform mole and fetus with normal karyotype: support of a separate entity. Obstet Gynecol 1991; 77: 868–874.
Golubovsky MD : Postzygotic diploidization of triploids as a source of unusual cases of mosaicism, chimerism and twinning. Hum Reprod 2003; 18: 236–242.
Niemann I, Bolund L, Sunde L : Twin pregnancies with diploid hydatidiform mole and co-existing normal fetus may originate from one oocyte. Hum Reprod 2008; 23: 2031–2035.
Surti U, Hill LM, Dunn J et al: Twin pregnancy with a chimeric androgenetic and biparental placenta in one twin displaying placental mesenchymal dysplasia phenotype. Prenat Diagn 2005; 25: 1048–1056.
Robinson WP, Lauzon JL, Innes AM et al: Origin and outcome of pregnancies affected by androgenetic/biparental chimerism. Hum Reprod 2007; 22: 1114–1122.
H’mida D, Gribaa M, Yacoubi T et al: Placental mesenchymal dysplasia with Beckwith-Wiedemann syndrome fetus in the context of biparental and androgenic cell lines. Placenta 2008; 29: 454–460.
Hoban PR, Heighway J, White GR et al: Genome-wide loss of maternal alleles in a nephrogenic rest and Wilms’ tumour from a BWS patient. Hum Genet 1995; 95: 651–656.
Giurgea I, Sanlaville D, Fournet JC et al: Congenital hyperinsulinism and mosaic abnormalities of the ploidy. J Med Genet 2006; 43: 248–254.
Wilson M, Peters G, Bennetts B et al: The clinical phenotype of mosaicism for genome-wide paternal uniparental disomy: two new reports. Am J Med Genet A 2008; 146A: 137–148.
Reed RC, Beischel L, Schoof J et al: Androgenetic/biparental mosaicism in an infant with hepatic mesenchymal hamartoma and placental mesenchymal dysplasia. Pediatr Dev Pathol 2008; 11: 377–383.
Romanelli V, Nevado J, Fraga M et al: Constitutional mosaic genome-wide uniparental disomy due to diploidisation: an unusual cancer-predisposing mechanism. J Med Genet 2011; 48: 212–216.
Morales C, Soler A, Badenas C et al: Reproductive consequences of genome-wide paternal uniparental disomy mosaicism: description of two cases with different mechanisms of origin and pregnancy outcomes. Fertil Steril 2009; 92: 393.e5–393.e9.
Hojberg KE, Aagaard J, Henriques U et al: Placental vascular malformation with mesenchymal hyperplasia and a localized chorioangioma. A rarity simulating partial mole. Pathol Res Pract 1994; 190: 808–813; discussion 814.
Palmer JR : Advances in the epidemiology of gestational trophoblastic disease. J Reprod Med 1994; 39: 155–162.
Genest DR, Berkowitz RS, Fisher RA : Gestational trophoblastic disease; in Tavassoli FA, Devilee P (eds): Health Organization Classification of Tumours. Pathology and Genetics. Tumors of the Breast and Female Genital Organs. Lyon: IARC Press, 2003, pp 250–256.
Zhao J, Xiang Y, Wan XR et al: Molecular genetic analyses of choriocarcinoma. Placenta 2009; 30: 816–820.
Vindelov LL, Christensen IJ, Nissen NI : A detergent-trypsin method for the preparation of nuclei for flow cytometric DNA analysis. Cytometry 1983; 3: 323–327.
Acknowledgements
We thank patients and staff at the departments of gynaecology and pathology in Jutland, Denmark for providing samples and clinical data. Laboratory technician Inger Juncker, cand.scient., PhD Lillian G Jensen, and cand.scient. Søren Pedersen are thanked for valuable help in the handling of data.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on European Journal of Human Genetics website
Rights and permissions
About this article
Cite this article
Sunde, L., Niemann, I., Hansen, E. et al. Mosaics and moles. Eur J Hum Genet 19, 1026–1031 (2011). https://doi.org/10.1038/ejhg.2011.93
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2011.93
Keywords
This article is cited by
-
Practical guidelines of the EOTTD for pathological and genetic diagnosis of hydatidiform moles
Virchows Archiv (2024)
-
Androgenetic/biparental mosaicism in a diploid mole-like conceptus: report of a case with triple paternal contribution
Virchows Archiv (2023)
-
Parental genomes segregate into distinct blastomeres during multipolar zygotic divisions leading to mixoploid and chimeric blastocysts
Genome Biology (2022)
-
Refined diagnosis of hydatidiform moles with p57 immunohistochemistry and molecular genotyping: updated analysis of a prospective series of 2217 cases
Modern Pathology (2021)
-
Molecular cytogenetic analysis of a hydatidiform mole with coexistent fetus: a case report
Journal of Medical Case Reports (2019)