Abstract
Population isolates have long been of interest to genetic epidemiologists because of their potential to increase power to detect disease-causing genetic variants. The Sorbs of Germany are considered as cultural and linguistic isolates and have recently been the focus of disease association mapping efforts. They are thought to have settled in their present location in eastern Germany after a westward migration from a largely Slavic-speaking territory during the Middle Ages. To examine Sorbian genetic diversity within the context of other European populations, we analyzed genotype data for over 30 000 autosomal single-nucleotide polymorphisms from over 200 Sorbs individuals. We compare the Sorbs with other European individuals, including samples from population isolates. Despite their geographical proximity to German speakers, the Sorbs showed greatest genetic similarity to Polish and Czech individuals, consistent with the linguistic proximity of Sorbian to other West Slavic languages. The Sorbs also showed evidence of subtle levels of genetic isolation in comparison with samples from non-isolated European populations. The level of genetic isolation was less than that observed for the Sardinians and French Basque, who were clear outliers on multiple measures of isolation. The finding of the Sorbs as only a minor genetic isolate demonstrates the need to genetically characterize putative population isolates, as they possess a wide range of levels of isolation because of their different demographic histories.
Similar content being viewed by others
Introduction
Population isolates have been favored as study populations for genetic mapping because of their unique demographic histories, which are typically characterized by long periods of isolation from other populations, and in some cases, increased levels of inbreeding and/or bottlenecks during their founding.1 As a result of their demography, population isolates are expected to have lower levels of genetic heterogeneity underlying disease traits, low levels of internal population structure and higher levels of linkage disequilibrium (LD), all of which aid mapping efforts.2 A further expected benefit is that the greater uniformity in environment and lifestyle, typically found in population isolates, allows for more facile matching of cases and controls and higher heritability of phenotypes (including disease traits).2
Whether many human population isolates in fact evidence substantially longer range LD and lower levels of genetic diversity, has been unclear. Early simulations and results argued for only modest differences between isolates and outbred populations.3, 4, 5 More recent empirical surveys show that isolates that have experienced a large founder event coupled with recent population growth, demonstrate higher overall levels of LD and fewer regions of very low LD (eg, Service et al6 Angius et al7). However, because each population isolate is unique, the potential effectiveness of any population isolate for mapping needs to be assessed by considering its specific demographic history and by undertaking empirical surveys of genetic variation.
The Sorbs, who reside in the Lusatia region of eastern Germany, are a population isolate defined by their use of a West Slavic language (Sorbian) in an area with a majority of Germanic speakers (Figure 1). The degree of isolation is more remarkable when one considers the more eastward distribution of Germanic territory and people before World War I. There are presently ∼25 000 Sorbian speakers, with 18 000 being speakers of Upper Sorbian (which is closer to the Czech language) residing in the federal state of Saxony and 7000 speakers of Lower Sorbian (closer to the Polish language) in the more northerly state of Brandenburg.8
One study of Sorbian populations describes their presence in present-day Germany as the result of an initial spread of numerous Slavic groups (collectively termed Polabians) westward from at least the 1st Century C.E. The geographical origins of these Polabians are essentially unknown, but various authors have placed it in present-day Ukraine, southern Belarus, parts of Russia, southern Poland and the Czech Republic. From the 9th century forward, eastward expansions of Germanic populations resulted in the displacement or absorption of virtually all Polabians, with the exception of the ancestors of modern Sorbs. Sorbs remain culturally distinct from their Germanic-speaking neighbors, preserving much of their Slavic language and traditional customs. A more detailed historical account with a bibliography is provided in the Supplementary Information.
Previous population genetic studies of Sorbian individuals have been limited to Y-chromosome surveys,9, 10, 11, 12 and have shown the greatest similarity of Sorbian individuals to be with West Slavic populations (with greatest affinity with Poles). Recently, the Sorbs have been successfully utilized in candidate gene and genome-wide association studies of height and metabolic syndrome traits.13, 14, 15, 16, 17 Although these studies are suggestive of the power of using the Sorbs for mapping, they do not make clear the extent to which Sorbs are a genetic isolate.
In this study, we examine patterns of genetic variation across Sorbian individuals in order to understand (a) the relationship of Sorbian individuals to other Europeans, which may offer unique insights into their demographic history, and (b) their potential status as a genetic isolate, which is of interest from a genetic epidemiology perspective. To accomplish this, we examine SNP variation found in Sorbian individuals within the context of data from a large number of previously genotyped outbred European individuals,18, 19, 20, 21 as well as newly genotyped German and Slovak individuals. In particular, we contrast genetic diversity in the Sorbs to that of French Basque and Sardinians, who have long been regarded as population isolates within Europe7, 22, 23 as well as the Orcadians, who are considered cultural isolates with less evidence for genetic isolation.24, 25
Subjects and methods
Data sets
Sample collection
A total of 996 Upper Sorbian speakers of the Catholic denomination were recruited from private practices in eight villages from Eastern Saxony. These individuals were initially thinned to a set consisting of 289 Sorbian individuals (124=male, 165=female), on the basis of the criteria that all four grandparents were described as Sorbian, pairwise identity-by-descent (IBD) was less than 0.15, SNP missingness was less than 7% per individual and single-nucleotide polymorphisms (SNPs) were successfully genotyped in at least 30% of individuals. We also conducted a more conservative filtering of these 289 individuals by a stricter IBD analysis that resulted in what would be considered a homogenous set of 178 unrelated Sorbs (see Supplementary Information). In addition, 10 and 15 individuals from Germany and Slovakia, respectively, were recruited at the University of Leipzig and Slovak Academy of Sciences. All studies were approved by the ethics committee of the University of Leipzig, and all subjects gave written informed consent before taking part in the study.
DNA extraction and genotyping
Genomic DNA was extracted using the QIAmp DNA Blood Midi Kit (Qiagen Inc., Valencia, CA, USA) according to the manufacturer's protocol. Genotyping was performed using the 500K Affymetrix and Affymetrix 6.0 GeneChip set (Affymetrix, Santa Clara, CA, USA). Probes were scanned by GeneChip Scanner 3000 7G and genotypes were determined with the GeneChip Genotyping Analysis Software (GTYPE) using the BRLMM or Birdseed algorithm. From this point on, we refer to this genotype data as the LPZ data set, in reference to the University of Leipzig, at which the work was completed.
Reference data set and merging
POPRES: genotype data for 1387 European individuals from the Population Reference Sample (POPRES) project,18 described in Novembre et al,19 were used for reference purposes. HGDP: genotype data for 140 European individuals from the Human Genome Diversity Project (HGDP) panel20, 21 were also used as reference data. Data set Merging and Quality control: We constructed three data set merges: POPRES/LPZ (30,587 SNPs), POPRES/HGDP (30,587 SNPs) and POPRES/LPZ/HGDP (8745 SNPS). The potential impact of batch effects within the LPZ data set was a major concern when merging with the POPRES and HGDP data sets (see Supplementary Figure 1). Therefore, we applied a very conservative approach, thinning to very high-quality SNP calls, indicated by low missingness values. These criteria suitably corrected for the initial difference between German individuals genotyped in the LPZ and HGDP data sets before filtering (see Supplementary Information for further details).
With regard to our power to identify population structure using a reduced set of SNPs, Patterson et al26 provide evidence that a threshold behavior exists for detecting population structure, such that if FST is greater than 1/√nm (where n equals the number of markers and m the total sample size), populations will be resolved in a principle components analysis (PCA). For n=30 000 (the number of SNPs in the POPRES/LPZ merge) and m=27 × 2=54 (where 27 is the median sample size across LPZ, POPRES and HGDP data sets), the minimum FST threshold would be 0.0008. To set this value in context, the FST between broad regions of Europe is 0.004,19 and the median FST across all pairwise comparisons we observe in our study is 0.0019 (see Supplementary Table 1). Thus, we expect that our conservative approach of filtering SNPs should not cost us very much in terms of detecting population structure.
Statistical analysis
Population structure analysis
PCA: all PCA was performed using smartpca26 under default parameters with 0 outlier detection iterations, unless otherwise stated. To control for the potential effect of uneven sampling and excess relatedness on PCA visualization,27 we implemented a ‘drop one in’ procedure for incorporating population isolates (see Supplementary Information for further details). PCA bootstrap analysis was performed as described by Novembre et al,19 Supplementary Material. Pairwise FST: pairwise population FST values were based on the θ statistic.28 A permutation test, using 10 000 replicates, was used for testing the relative significance of pairs of distances. Ancestry estimation: maximum likelihood estimation of individual ancestries was performed using the software Admixture (http://www.genetics.ucla.edu/software/admixture/),29 using default values (see Supplementary Information for further details).
Measures of inbreeding
Inbreeding coefficient, F, and runs of homozygosity (ROH): calculation of the inbreeding coefficient, F, and analysis of ROH were performed using PLINK routines (http://pngu.mgh.harvard.edu/purcell/plink/; see Supplementary Information for further details). Linkage disequilibrium Decay (LD) analysis: LD was quantified using both the genotypic- and haplotypic-based r2 statistic for the POPRES/LPZ and POPRES/HGDP data sets (before any LD filtering). We followed the resampling procedure of Jakobsson et al30 to control for uneven sample size (see Supplementary Information for further details). Haplotype phasing was performed using fastPHASE version 1.2 with a K-value (number of haplotypes clusters considered) of 20 (as utilized in Pickrell et al31), and the use of subpopulation labels based on country of origin for POPRES samples and ethnic identity for HGDP and LPZ samples if available. All other parameters were run under default values.
Results
Sorbs in the context of broader European genetic diversity
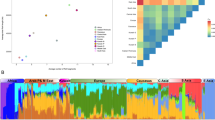
To assess how the Sorbs fit within a general background of spatial structure among European populations,19 we conducted PCA on the merged LPZ/POPRES data set. The median position of all Sorbs (Figure 2a) is roughly representative of their geographical location, that is, west of Poles, north of Czechs and east of Germans. The bootstrap distribution of the median PC1–PC2 position of the Sorbs overlaps that of the Poles, demonstrating their genetic similarity, but was distinct from both the Germans and Czechs (Figure 2c). The median position for the LPZ Slovakian individuals was also located in approximately the expected position based on geography (suggesting the outlier position of the single POPRES Slovakian individual in Novembre et al19 was a sampling artifact).
(a) PCA analysis using POPRES/LPZ merge. Circled position indicates median coordinate value for Sorbs. (b) Same as a, except using POPRES/HGDP merge. (c) Bootstrap analysis via median coordinate values of POPRES/LPZ merge based on sample's country of origin. (d) Same as c, except using POPRES/HGDP merge. PCA1 is on y axis and PCA2 is on x axis for all plots. For (a, b) an appending asterisk indicates that this individual (and sample median position) is from the Leipzig data set. For (c, d) an appending asterisk indicates that this individual (and sample median position) is from the HGDP data set.
PCA analysis using the POPRES/HGDP merge (Figure 2b) showed both the Basque and Sardinians to be highly differentiated from all other Europeans, with clearly discrete median bootstrap distributions (Figure 2d). Interestingly, five Italians from the POPRES data set that were outliers in Novembre et al,19 clustered tightly with the HGDP Sardinians. In addition, the coordinates of HGDP Northern Italians and Tuscans reflect their relative geographical positions when compared with the more general set of POPRES Italians. The Orcadians are almost completely indistinguishable from individuals from the United Kingdom and Ireland.
Pairwise FST values were calculated between all populations with a sample size of 10 or greater (Supplementary Table 1) using the POPRES/LPZ/HGDP merge (a total of 26 populations using 8745 SNPs). The Sorbs were generally found to be closer to Slavic- than Germanic-speaking population. Examining these distances in more detail showed the Sorbs to be closer to Czechs than Poles (Table 1); but this difference was not statistically significant when assessed by permutation (P=0.280). However, the FST values between the Sorbs and both Czechs and Poles were significantly smaller (P<0.05) than that between the Sorbs and any other population, including both German groups (P<0.001). Interestingly, the French Basque were significantly closer to Spanish POPRES individuals than either POPRES or HGDP French individuals (P<0.001). The magnitude of the FST was also informative: the Sorbs were much closer genetically to their geographical neighbors (eg, FST of Sorbs vs Czech=0.0008) than the French Basque and Sardinians were to their geographical neighbors (eg, FST of Basque vs French=0.0055, Sardinians vs Italians=0.0053) and other populations in general.
We estimated individual ancestry components in an unsupervised (ie, no parental groups were assumed) admixture model, using the maximum likelihood approach implemented in Admixture.29 The Sorbs (n=178) were analyzed alongside individuals from the following populations: POPRES Germany (DE, n=71), Poland (PL, n=22), Czech Republic (CZ, n=11), Serbia and Montenegro (YG, n=41), and LPZ Germany (LPZ-DE, n=10) and Slovakia (LPZ-SK, n=15). These populations were included alongside the Sorbs, as they were considered as potential source populations based either on geography (eg Germans) or historical hypotheses (eg, Sorbs potentially share recent common ancestry with Polish, Czech or Serbian individuals). The vast majority of individuals, including Sorbs, evidenced substantial ancestry from almost all K inferred clusters (Supplementary Figure 2), regardless of the K value used, reflecting a poor fit of the admixture model. Using the cross-validation (CV) approach implemented in Admixture, we calculate the highest CV accuracy for values of K=1, with some overlap with K=2 (Supplementary Figure 3a), suggesting weak evidence for structure in this data. When ancestry components were averaged within populations (Supplementary Figure 4), the Sorbs were somewhat more distinct from other populations. For K=2, 3 and 4, the Sorbs showed greatest similarity to Poles, followed by Czechs, Slovaks and the two German populations. The Sorbs were most different to Serbia and Montenegro, likely reflecting the considerable geographical distance between the two populations. The above results were in stark contrast to that of the Sardinian and French Basque (assessed using a separate POPRES/HGDP merge), who are both clearly differentiated from neighboring European populations (Supplementary Figure 4, Supplementary Figure 5) for values of K=3 and above (the CV accuracy was highest for K=2, with substantial overlap with K=3 (Supplementary Figure 3b), a likely result of the emergence of Sardinian and Basque clusters), whereas the Orcadians showed only moderate evidence of differentiation.
Sorbs as a genetic isolate
We next investigated measures of genetic diversity in the Sorbs that might reflect a history of relative isolation, elevated levels of inbreeding and/or recent historical bottlenecks. We compare the values observed with those for the French Basque, Sardinian and Orcadians as well as other European reference populations.
As may be expected, the French Basque and Sardinians show evidence of substantial genetic isolation. The Sardinians, closely followed by the Basque, had extremely high pairwise FST values compared with all other populations, with the minimum FST with their geographic neighbors being 0.0053 (SD vs IT) and 0.0047 (BSQ vs ES), respectively (Supplementary Table 1). These minimum FST values were far higher than any other pairwise comparison between geographic neighbors, which ranged from −0.0014 to –0.0017 (ignoring any comparisons involving LPZ_DE, which tended to produce much lower values than expected). The MAF spectra (Supplementary Figure 6), although highly distorted because of SNP ascertainment, also show the Sardinians and Basques to have a noticeable excess of monomorphic SNPs. This excess suggests that some SNPs that are polymorphic in Europe may have been driven to extinction/fixation at a higher rate or never existed at all in these populations, consistent with genetic isolation. They were also both clear outliers with regard to the number of homozygous segments detected (Figure 3c). A plot of LD decay based on genotypic r2 shows the Basque to have slightly elevated LD (Figure 4b) that increases when using phased haplotype data (Figure 4d). Sardinians only demonstrated elevated LD using phased data, and were not comparable to the level observed in the Basque.
Box plots based on ROH analysis with samples grouped within all populations with n≥10. (a) Total number of homozygous segments within POPRES/LPZ merge, (b) average length of homozygous segments within POPRES/LPZ merge, (c) total number of homozygous segments within POPRES/HGDP merge and (d) average length of homozygous segments within POPRES/HGDP merge. Populations ordered from lowest to highest median values. Sorbs, French Basques, Sardinians and Orcadians indicated by yellow, red, blue and green coloring respectively.
Analysis of the Sorbs and Orcadians did indicate some subtle signals of genetic isolation, albeit not to the same extent as the Basque and Sardinians. They had the fourth and fifth highest average pairwise FST values as well as the fifth and joint third highest minimum pairwise FST values, respectively (again ignoring LPZ_DE comparisons), although the magnitude of both these measures was approximately half of those seen in the Basques and Sardinians (Supplementary Table 1). The HGDP Russians were third and joint third in the average and minimum pairwise FST ranking, respectively, probably reflecting their relatively large geographical distance from other European populations in the POPRES, which has few populations from Eastern Europe. Similarly, the Sorbs and Orcadians possessed the third and fourth highest number of monomorphic SNPs (Supplementary Figure 6). The Sorbs were relatively unremarkable with regard to ROH (Figures 3a and b), but the Orcadians had the highest median average length of individual ROH segments (Figure 3d), suggesting a very recent history of inbreeding. Although not directly comparable to the other population isolates, the Sorbs show a consistent but only marginally elevated level of LD using genotypic data (Figure 4a) that is enhanced by phased data (Figure 4c). Orcadians appear to have a similar level of LD to Sardinians (Figures 4b and d).
All four population isolates were unremarkable with regard to their distribution of inbreeding coefficient F-values (Supplementary Figure 7). Interestingly, the vast majority of populations, including the putative population isolates, demonstrated negative median F-values.
Discussion
For most measures of isolation that we considered, the Sorbs demonstrated values the direction of which is indicative of isolation relative to other non-isolated European populations. At the same time, the magnitude of these values was small, and thus the Sorbs appear very modestly isolated in comparison with the French Basque and Sardinians, who consistently showed substantial signatures of isolation. The Sorbs are much like the Orcadians: for many measures they are at or toward the extreme of the observed distribution across European reference populations but they are never clear outliers.
One possible explanation for this effect is that the historical evidence for the timing of the establishment of the Sorbs (∼9th century AD) suggests a relatively recent origin in contrast to the potentially much more ancient Sardinian (at least 5000 years32) and Basque populations (at least 18 000 years33). Topography may also be relevant in the fact that Sardinians are isolated on an island and Basque live in a mountainous region. The Sorbs are separated from their neighbors predominantly because of linguistic and religious differences, which may be weaker forces for maintaining isolation than topographical barriers. Indeed, many medieval scholars have concluded that much of European demographic history between the Middle Ages and the present has been very fluid and that the long-term demographic continuity of particular ethnic groups, such as the Sorbs, is questionable.34 In this context, it is perhaps less surprising that our sample of Sorb individuals appears only modestly isolated from other populations in the region.
As previously observed using the Y chromosome,9, 10, 11, 12 the data presented in this study show that the Sorbs have greatest genetic similarity to western Slavic-speaking populations, despite being surrounded by predominantly Germanic speakers. This result contrasts the broad-scale pattern in Europe, in which genetic variation is strongly driven by geographical proximity.19, 35 More specifically, the Sorbs showed greatest affinity to Polish and Czech populations. Although PCA visually seems to suggest that our Sorb sample is closer to Poles than Czech, there is no significant difference in pairwise FST values.
How well the Sorbs’ current genetic similarity to western Slavic speakers reflects their ethnogenesis is unclear. A major unknown fact of Sorbian history is the start site of the migration of Slavic Polabians, with hypotheses ranging from more western origins in Poland through more eastern origins in Ukraine and Russia. Sorbs may appear similar to western Slavic speakers because of recent gene flow or recent population divergence from western Slavic speakers. We explored fitting formal demographic models representing descent from a western vs eastern Slavic population within an Approximate Bayesian Computation framework, but found that we could not reliably distinguish the models (controlling for SNP ascertainment bias seems to have limited power).
For the purposes of genetic epidemiology, our study shows that Sorbs evidence only modest signs of isolation, and so potential benefits for mapping would reside in other factors than isolation. Given their shared culture and geographic proximity, they may have a relatively consistent environment across individuals, which would increase power for investigating traits that would otherwise have large environmental components or genotype–by–environment interactions. In addition, the Sorbs are genetically similar to Slavic-speaking Europeans, who are understudied in genome-wide association studies relative to western European populations, and thus may contain rare disease variants that were previously not found.
One caution regarding our results is that the geographical origins of our reference populations are crudely characterized only by country and thus may not be random samples. If many of the Germans in the POPRES data are western German samples, this may inflate the apparent differences we observe between Germans and Sorbs. The LPZ Germans contained two individuals from Eastern Germany who do appear closer to the Sorbs, suggesting that population structure within countries is a valid concern. Certainly, a tighter and denser sampling of German, Polish and Czech individuals from regions surrounding the Sorbian territories would be ideal for confirming or refuting the results found in this study. Another caveat is that the modest elevation in LD for any of our population isolates may be due to a lack of power. Some studies6, 7, 36 have found elevated LD in isolates by using much larger sample sizes (eg, >100 individuals), whereas to control for uneven sampling our sample size was effectively reduced to 10 individuals. Nevertheless, other studies also show only subtle differences between putative isolates and other populations.37
Our results are particularly interesting in light of recent studies in which the Basques have showed no or little evidence of differentiation from neighboring European populations.38, 39 The discrepancy between these and our own study may be a result of different samples (Laayouni et al39 used Spanish rather than French Basque and much more well-defined comparative populations). However, the large difference in the signals of isolation between the two studies suggests that the 120 Ancestry Informative Markers used by Laayouni et al39 lack sufficient power. Indeed, a recent study,40 with a similar level of power to our own (60 000 SNPs), demonstrates evidence of Basque genetic isolation.
We encourage further studies on other population isolates as each study improves our understanding of patterns of genetic variation at spatially fine scales and helps untangle how historical relationships among human groups have structured this variation. For example, previous studies on Iceland41 and parts of rural Europe42 have also shown how high-throughput SNP approaches can be used to elucidate genetic relationships even at very fine geographical scales. Resequencing data will be especially powerful and revealing given the much smaller ascertainment biases associated with analyzing sequence variation.
References
Heutink P, Oostra BA : Gene finding in genetically isolated populations. Hum Mol Genet 2002; 11: 2507–2515.
Kristiansson K, Naukkarinen J, Peltonen L : Isolated populations and complex disease gene identification. Genome Biol 2008; 9: 109.
Kruglyak L : Prospects for whole-genome linkage disequilibrium mapping of common disease genes. Nat Genet 1999; 22: 139–144.
Eaves IA, Merriman TR, Barber RA et al: The genetically isolated populations of Finland and sardinia may not be a panacea for linkage disequilibrium mapping of common disease genes. Nat Genet 2000; 25: 320–323.
Dunning AM, Durocher F, Healey CS et al: The extent of linkage disequilibrium in four populations with distinct demographic histories. Am J Hum Genet 2000; 67: 1544–1554.
Service S, DeYoung J, Karayiorgou M et al: Magnitude and distribution of linkage disequilibrium in population isolates and implications for genome-wide association studies. Nat Genet 2006; 38: 556–560.
Angius A, Hyland FC, Persico I et al: Patterns of linkage disequilibrium between SNPs in a Sardinian population isolate and the selection of markers for association studies. Hum Hered 2008; 65: 9–22.
Ethnologue: Languages of the World; Vol. Sixteenth. SIL International Online, http://www.ethnologue.com/.
Behar DM, Thomas MG, Skorecki K et al: Multiple origins of Ashkenazi levites: y chromosome evidence for both near Eastern and European ancestries. Am J Hum Genet 2003; 73: 768–779.
Rodig H, Grum M, Grimmecke HD : Population study and evaluation of 20 Y-chromosome STR loci in Germans. Int J Legal Med 2007; 121: 24–27.
Immel UD, Krawczak M, Udolph J et al: Y-chromosomal STR haplotype analysis reveals surname-associated strata in the East-German population. Eur J Hum Genet 2006; 14: 577–582.
Krawczak M, Lu TT, Willuweit S, Roewer L : Genetic diversity in the German population. in: Cooper D, Kehrer-Sawatzki H (eds): Handbook of Human Molecular Evolution. Wiley: Chichester, 2008, Vol 12, pp 451–456.
Bottcher Y, Unbehauen H, Kloting N et al: Adipose tissue expression and genetic variants of the bone morphogenetic protein receptor 1A gene (BMPR1A) are associated with human obesity. Diabetes 2009; 58: 2119–2128.
Schleinitz D, Carmienke S, Bottcher Y et al: Role of genetic variation in the cannabinoid type 1 receptor gene (CNR1) in the pathophysiology of human obesity. Pharmacogenomics 2010; 11: 693–702.
Tonjes A, Koriath M, Schleinitz D et al: Genetic variation in GPR133 is associated with height: genome wide association study in the self-contained population of Sorbs. Hum Mol Genet 2009; 18: 4662–4668.
Tonjes A, Zeggini E, Kovacs P et al: Association of FTO variants with BMI and fat mass in the self-contained population of Sorbs in Germany. Eur J Hum Genet 2010; 18: 104–110.
Hoffmann K, Planitz C, Ruschendorf F et al: A novel locus for arterial hypertension on chromosome 1p36 maps to a metabolic syndrome trait cluster in the Sorbs, a Slavic population isolate in Germany. J Hypertens 2009; 27: 983–990.
Nelson MR, Bryc K, King KS et al: The population reference sample, POPRES: a resource for population, disease, and pharmacological genetics research. Am J Hum Genet 2008; 83: 347–358.
Novembre J, Johnson T, Bryc K et al: Genes mirror geography within Europe. Nature 2008.
Cann HM, de Toma C, Cazes L et al: A human genome diversity cell line panel. Science 2002; 296: 261–262.
Li JZ, Absher DM, Tang H et al: Worldwide human relationships inferred from genome-wide patterns of variation. Science 2008; 319: 1100–1104.
Arcos-Burgos M, Muenke M : Genetics of population isolates. Clin Genet 2002; 61: 233–247.
Cavalli-Sforza LL, Menozzi P, Piazza A : The History and Geography of Human Genes. New Jersey: Princeton University Press, 1994.
Roberts DF : Who are the Orcadians? Anthropol Anz 1986; 44: 93–104.
Roberts DF : Genetic affinities of the Shetland islanders. Ann Hum Biol 1990; 17: 121–132.
Patterson N, Price AL, Reich D : Population structure and eigenanalysis. PLoS Genet 2006; 2: e190.
McVean G : A genealogical interpretation of principal components analysis. PLoS Genet 2009; 5: e1000686.
Weir BS, Cockerham CC : Estimating F-statistics for the analysis of population structure. Evolution 1984; 38: 1358–1370.
Alexander DH, Novembre J, Lange K : Fast model-based estimation of ancestry in unrelated individuals. Genome Res 2009; 19: 1655–1664.
Jakobsson M, Scholz SW, Scheet P et al: Genotype, haplotype and copy-number variation in worldwide human populations. Nature 2008; 451: 998–1003.
Pickrell JK, Coop G, Novembre J et al: Signals of recent positive selection in a worldwide sample of human populations. Genome Res 2009; 19: 826–837.
Passarino G, Underhill PA, Cavalli-Sforza LL et al: Y chromosome binary markers to study the high prevalence of males in Sardinian centenarians and the genetic structure of the Sardinian population. Hum Hered 2001; 52: 136–139.
Calafell F, Bertranpetit J : Principal component analysis of gene frequencies and the origin of Basques. Am J Phys Anthropol 1994; 93: 201–215.
Geary PJ : The myth of nations: the medieval origins of Europe. Princeton: Princeton University Press, 2002.
Lao O, Lu TT, Nothnagel M et al: Correlation between genetic and geographic structure in Europe. Curr Biol 2008; 18: 1241–1248.
Tenesa A, Wright AF, Knott SA et al: Extent of linkage disequilibrium in a Sardinian sub-isolate: sampling and methodological considerations. Hum Mol Genet 2004; 13: 25–33.
Bosch E, Laayouni H, Morcillo-Suarez C et al: Decay of linkage disequilibrium within genes across HGDP-CEPH human samples: most population isolates do not show increased LD. BMC Genomics 2009; 10: 338.
Garagnani P, Laayouni H, Gonzalez-Neira A et al: Isolated populations as treasure troves in genetic epidemiology: the case of the Basques. Eur J Hum Genet 2009; 17: 1490–1494.
Laayouni H, Calafell F, Bertranpetit J : A genome-wide survey does not show the genetic distinctiveness of Basques. Hum Genet 2010; 127: 455–458.
Rodriguez-Ezpeleta N, varez-Busto J, Imaz L et al: High-density SNP genotyping detects homogeneity of Spanish and French Basques, and confirms their genomic distinctiveness from other European populations. Hum Genet 2010; 128: 113–117.
Price AL, Helgason A, Palsson S et al: The impact of divergence time on the nature of population structure: an example from Iceland. PLoS Genet 2009; 5: e1000505.
O’Dushlaine C, McQuillan R, Weale ME et al: Genes predict village of origin in rural Europe. Eur J Hum Genet 2010; 18: 1269–1270.
Acknowledgements
The authors thank all sample donors, Knut Krohn (Microarray Core Facility of the Interdisciplinary Centre for Clinical Research, University of Leipzig) for the genotyping support, and Mark I McCarthy and Nigel William Rayner for analytical and bioinformatics support. Financial support was received from the Andrew W Mellon Foundation (KV), Searle Scholars Program (JN), German Research Council (KFO-152), IZKF (B27) and the German Diabetes Association. AG and MSc were funded by the Leipzig Interdisciplinary Research Cluster of Genetic Factors, Clinical Phenotypes and Environment (LIFE Center, University of Leipzig). LIFE is funded by means of the European Union, by the European Regional Development Fund (ERFD) and by means of the Free State of Saxony within the framework of the excellence initiative. IK and DG were funded by grants from the Slovak Diabetes Association and the ERDF (Transendogen/26240220051).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Veeramah, K., Tönjes, A., Kovacs, P. et al. Genetic variation in the Sorbs of eastern Germany in the context of broader European genetic diversity. Eur J Hum Genet 19, 995–1001 (2011). https://doi.org/10.1038/ejhg.2011.65
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2011.65
Keywords
This article is cited by
-
Genome-wide meta-analysis of phytosterols reveals five novel loci and a detrimental effect on coronary atherosclerosis
Nature Communications (2022)
-
Genetically programmed changes in transcription of the novel progranulin regulator
Journal of Molecular Medicine (2020)
-
Metabolic effects of genetic variation in the human REPIN1 gene
International Journal of Obesity (2019)
-
On the impact of relatedness on SNP association analysis
BMC Genetics (2017)
-
Overcoming the dichotomy between open and isolated populations using genomic data from a large European dataset
Scientific Reports (2017)