Abstract
Mutation of the LARGE gene is the rarest of the six known genetic causes of α-dystroglycanopathy. We report further a family with MDC1D due to a complex genomic rearrangement that was not apparent on standard sequencing of LARGE. Two sisters in a consanguineous family had moderate mental retardation and cerebellar malformations, together with dystrophic changes and markedly reduced α-dystroglycan glycosylation staining on muscle biopsy. There was homozygous linkage to the LARGE locus but sequencing of LARGE coding regions was normal. Analysis of LARGE cDNA showed an abnormal sequence inserted between exons 10 and 11, in most of the transcripts, predicted to introduce a premature stop codon. The abnormal sequence mapped to a spliced EST (DA935254) of unknown function, normally located at 100 kb centromeric of LARGE on chromosome 22q12.3. Quantitative PCR analysis of the EST and adjacent regions showed twice the normal copy number in patients’ genomic DNA samples, consistent with a large intra-chromosomal duplication inserted into intron 10 of LARGE in a homozygous state. This insertion was associated with deletion of a central region of intron 10, but the exact break points of the deletion/duplication were not found, suggesting that an even more complex rearrangement may have occurred. The exact function of LARGE, a golgi protein, remains uncertain. POMT and POMGnT enzyme activities were normal in patients’ lymphoblast cells, suggesting that defects in LARGE do not affect the initiation of O-mannosyl glycans.
Similar content being viewed by others
Introduction
Alpha-dystroglycan (αDG) is a highly glycosylated cell surface protein that has important roles in neuronal cell migration and cell-to-extracellular matrix interactions in muscle. Abnormality in the glycosylation of αDG is the hallmark histological abnormality and the likely pathogenic mechanism for a group of congenital muscular dystrophies (CMD), collectively called the ‘α-dystroglycanopathies’.1 At present, six genes have been linked to this common clinicopathological presentation (POMT1, POMT2, POMGNT1, FKTN, FKRP and LARGE). All of these encode confirmed or putative enzymes that are thought to function in the O-mannosyl glycosylation pathway. Abnormal αDG glycosylation correlates with an increasingly severe pattern of tissue effects that extend from an adult-onset limb-girdle muscular dystrophy with normal intelligence (LGMD type 2I) to severe congenital brain and eye malformations (Walker–Warburg syndrome, WWS).1 The LARGE gene became a candidate for human CMD after the discovery of the myodystrophic (myd) mouse that has a spontaneous null mutation in Large because of a genomic deletion involving exons 5 to 7.2, 3, 4 Another spontaneous murine mutant, named veils (vls) also results from an intragenic deletion leading to loss of exons 3 to 5 and premature protein termination.5 The two strains share typical cerebral, ocular and muscular changes of an α-dystroglycanopathy. MDC1D due to mutations in LARGE is the rarest of the α-dystroglycanopathies identified to date, with only three confirmed families previously described.6, 7, 8 Affected children have had typical neurological and muscle abnormalities associated with the α-dystroglycanopathies, but with very different severities; one family had mild muscle-eye-brain disease (MEB) and the other two had typical WWS. Another WWS patient has been reported with a single heterozygous nonsense mutation in LARGE.9 Confirmation that this patient has CMD due to LARGE awaits the discovery of a second mutation. In this paper, we describe the fourth confirmed family with MDC1D due to a homozygous LARGE mutation, in which two sisters from consanguineous parents have moderate mental retardation, cerebellar and pontine hypoplasia, and persistent white matter abnormalities on brain magnetic resonance imaging (MRI), features consistent with mild MEB. We identified an unusual genetic mechanism for LARGE gene disruption, namely the splicing of an abnormal sequence into the LARGE mRNA transcript likely due to a duplication/deletion event involving intron 10. This report extends our knowledge of the clinical phenotype associated with LARGE, and is the second family in which disease results from a large-scale gene rearrangement.
The POMT1, POMT2 and POMGNT1 genes encode enzymes that together mediate the first two steps in the O-mannosyl glycosylation pathway. A recent study suggests that LARGE participates in post-phosphoryl glycosylation of a novel phosphorylated O-mannosyl glycan on the mucin-like domain of recombinant αDG, which is required for laminin binding.10 Evidence from cell culture and a fukutin transgenic mouse suggests that fukutin physically interacts with POMGnT1 and can cause reduced POMGnT1 activity when abnormal.11 To investigate whether LARGE may also function by influencing the first steps of the O-mannosyl pathway, we assessed the activity levels of POMT and POMGnT1 in cultured lymphoblasts from one of our patients.
Materials and methods
Case reports
The two affected girls are the only children of first-cousin parents from Lebanon. There is no other family history of neuromuscular disease. Both children were born full-term after uneventful pregnancies with normal foetal movements and deliveries. Birth weights, lengths and head circumferences were within normal limits.
Patient 1: The older sibling first presented to medical attention at an age of 2.5 months with hypotonia and regurgitation. Acquisition of all motor skills was delayed. She sat at an age of 9 months, stood unsupported at an age of 2.5 years and at an age of 8 years, she could climb stairs with difficulty but could not run. Her full-scale IQ was 61. On examination at an age of 8 years, she weighed 24 kg (50th centile), her height was 122.5 cm (35th centile) and the occipitofrontal circumference (OFC) was 53 cm (75th centile). Her speech was restricted to dysarthric simple words, and she followed simple commands. She had a myopathic face with an open-mouth posture and mild tongue hypertrophy. There was generalised muscle hypertrophy, most prominent in the calves, thighs and the shoulder girdle. This contrasted with moderate proximal lower limb weakness that manifested as a waddling gait and a positive Gowers’ manoeuvre. Upper limb strength was within normal limits. There were no joint contractures, except for mild tightness of the Achilles’ tendons, associated with toe walking. The only abnormalities found on eye assessment were mild myopia and strabismus. There was no history of seizures. Cardiac and respiratory examinations were normal. Brain MRI performed at ages 3 ½ and 6 years showed marked cerebellar atrophy (particularly affecting the vermis) and cerebellar cysts (Figure 1). There was mild generalised cerebral and pontine atrophy, dilatation of all ventricular spaces and pachygyria of the frontal lobes (Figure 1). Diffuse symmetrical high signal abnormalities were seen within the periventricular and deep white matter of both cerebral hemispheres, with sparing of the subcortical U fibres. These hyperintensities were unchanged at the age of 6 years. A generalised skeletal X-ray survey, abdominal ultrasound, echocardiogram, electroencephalogram and auditory-evoked potentials with brainstem responses were all normal. Creatine kinase (CK) levels were 30 times higher than the upper limit of normal.
Brain MRI images obtained from Patient 1 performed at an age of 3.5 years. (a) Coronal T1-weighted image showing cerebellar and vermis hypoplasia (white arrows), cerebellar cysts (black arrow), generalised white matter atrophy and increased CSF spaces. (b) Transverse T2-weighted image showing abnormal high white matter signal and dilated lateral ventricles. (c) Sagittal T1-weighted image showing cerebellar (white arrow) and pontine hypoplasia.
Patient 2: The younger sister followed a clinical course similar to her sister, but was less delayed in her motor and cognitive skills. She stood unsupported at an age of 16 months and walked independently at an age of 2 years. On examination at an age of 4 ½ years, her weight was 15 kg (25th centile), height was 106 cm (75th centile) and OFC was 51 cm (50th centile). There was generalised hypotonia, firm hypertrophic calves and a mild equinovarus foot position. Mild myopia was the only abnormality observed on eye examination. Myopathic features were noted on EMG analysis, whereas nerve conduction studies were normal. Brain MRI performed at 2 years showed diffuse high signal white matter abnormalities, mainly in the frontal and parieto–occipital regions, dilated ventricular spaces and marked cerebellar hypoplasia. CK levels were 60 times above the upper limit of normal.
Muscle biopsy findings
Muscle and skin biopsy samples were taken from Patient 1 from the right gastrocnemius muscle. Standard histochemical analysis revealed a dystrophic pattern characterised by fibre size variation, markedly increased connective tissue, internalised nuclei and scattered necrotic fibres. Immunohistochemistry (IHC) for αDG showed markedly reduced staining using VIA4-1 (Millipore, Billerica, MA, USA), an antibody that recognises a glycosylated epitope. Laminin-α2 expression was mildly reduced by IHC using antibodies that recognize both the 80 kDa (clone 5H2; Millipore) and the 300 kDa (NCL-merosin; Novocastra Labs, Newcastle, UK) fragments. Staining for β-dystroglycan (βDG), dystrophin, collagen VI and sarcoglycans (α and γ) was normal.
Western blot analysis
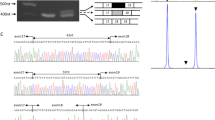
Western blotting was used to investigate the degree of abnormal αDG glycosylation using a skin fibroblast cell line obtained from Patient 1, as frozen muscle was not available. Fibroblasts were maintained at 37°C and 5% CO2 in DMEM medium plus 20% foetal bovine serum and 0.5% penicillin-streptomycin (Gibco, Invitrogen Corp., Carlsbad, CA, USA). Cultured cells were lysed, and glycoproteins from cell homogenates were enriched with WGA lectin as previously described.11 We performed immunoblots (for αDG and βDG) and laminin ligand overlay assays using polyvinylidene difluoride membranes, and blots were developed by horseradish peroxidase-enhanced chemiluminescence (Pierce, Thermo Fisher Scientific, Rockford, IL, USA).12 The monoclonal antibodies IIH-6 and VIA4-1, which are specific for functionally glycosylated αDG,13 showed no detectable binding affinity to αDG obtained from patient fibroblast cells, even though βDG (monoclonal antibody 7D11) was normally present, consistent with a glycosylation defect in αDG (Figure 2). This defect in αDG glycosylation also resulted in loss of laminin-1 ligand binding (Figure 2).
Western blot analysis of α-dystroglycan. Immunoblot analysis of WGA-enriched homogenates from fibroblasts obtained from Patient 1 and a healthy individual (control). Absent staining of antibodies IIH6 and VIA4 that bind to glycosyl epitopes on α-dystroglycan (α-DG) and a laminin-1 (Lam) overlay assay show abnormalities in αDG glycosylation in patient fibroblasts that is associated with loss of laminin-1 protein binding. Beta-dystroglycan (β-DG) is used as a loading control.
Genetic analysis
We performed linkage analysis using microsatellite markers to the five α-dystroglycanopathy loci known at the time (POMGNT1, FKRP, POMT1, POMT2 and FKTN) and the DAG1 gene that encodes DG, but we identified no homozygous linkage. A genome-wide linkage study using 400 microsatellite markers showed potential linkage to the newly identified LARGE locus and analysis of further markers supported homozygous linkage (Figure 3). We sequenced the coding regions of LARGE bi-directionally by standard techniques. Oligonucleotide primers, at least 80 base pairs (bp) outside the exons, were chosen for PCR amplification and sequencing on ABI 3730 capillary electrophoresis system (Applied Biosystems, Life Technologies Corp., Carlsbad, CA, USA), but no abnormalities were found. Subsequently, mRNA was extracted from cultured EBV-transformed lymphoblasts obtained from Patient 1 (Trizol method), cDNA was generated using oligodT Superscript II First-Strand Synthesis (Invitrogen, Carlsbad, CA, USA) and the LARGE gene transcript was sequenced using 14 overlapping primer pairs. Two abnormally large PCR products, together with a faint product of the expected size, were obtained for primers that spanned between exons 9 and 12 (Figure 4a). Sequencing revealed that most mRNA transcripts contained an abnormal 197 bp sequence inserted between exons 10 and 11. Low levels of a 171 bp insertion (identical to the 197 bp sequence except lacking 26 bases from the 3′ end) and a normally spliced transcript were also obtained. Identical splicing abnormalities were also seen in mRNA from cultured patient fibroblast cells, except a greater proportion of transcripts showed normal splicing (Figure 4a). The abnormal sequences mapped to a part of a spliced EST (GenBank accession no: DA935254) of unknown function, originally identified in a human splenic cDNA library,14 normally located at 100 kb centromeric to the LARGE gene on chromosome 22q12.3 (Figure 5). The presence of the two most abundant abnormally inserted sequences are predicted to immediately introduce a premature stop codon in the LARGE mRNA transcript, resulting in truncation of the LARGE protein mid-way through translation (Figure 4b). Analysis of the inserted EST sequence identified potential splice acceptor and donor sites for both the larger and smaller insertions that likely mediate the abnormal splicing (Supplementary E, Figure 1). Archived frozen muscle was no longer available to verify the abnormalities in LARGE splicing in muscle.
Analysis of the LARGE gene insertion in genomic and cDNA. (a) Agarose gel showing the PCR products from patient (P) and control (C) cDNA generated from cultured lymphoblasts (Lymph) and fibroblasts (Fib) using primers that span from exons 9 to 12 of LARGE (Supplementary E, Table 1). In patient cDNA sample, two abnormally high bands predominate that correspond to insertions of abnormal DNA sequences between exons 10 and 11. A faint band of the normal size is also seen (arrow). (b) Diagram showing the amino acid sequence that is coded by normal and patient cDNA around the exon 10/11 boundary. The abnormal insertion found in patient cDNA results in the introduction of a premature STOP codon, predicted to cause protein termination mid-way through the translation of the LARGE protein. (c) Results from qPCR analysis of genomic DNA (gDNA) for EST DA935254 and the surrounding chromosomal region. gDNA concentrations between samples were adjusted using qPCR results from a distant gene (PTPLA) and results were scaled so that control samples had a mean copy number of 2 at each point (data not shown). In patients’ gDNA sample, qPCR analysis of the EST sequence and for five regions, located at variable distances upstream and downstream, shows twice the expected copy number consistent with homozygous duplication of this region. Genomic copy number returns to normal for markers 5b and 3b in the patient gDNA samples, and therefore these markers define the maximum size of the duplication. Analysis of gDNA from both parents shows that both had three copies of the duplicated region, consistent with each having the duplication in a heterozygous state. All results are shown relative to the reverse gDNA strand.
Diagram of the insertion/deletion mutation involving intron 10 of the LARGE gene. The EST DA935254, normally located at 100 kb centromeric of LARGE, is duplicated together with ∼40 kb of flanking DNA and likely inserted into intron 10 of LARGE. Numerous repetitive elements, which may have contributed to the large intra-chromosomal rearrangement, are present in both intron 10 and flanking the EST, including SINEs (green), LINEs (red) and LTRs (blue). bp=base pairs.
To investigate whether the abnormal LARGE splicing arose because of a chromosomal duplication, we performed quantitative PCR (qPCR) analysis of the EST sequence obtained from patients’ genomic DNA (gDNA) using the LightCycler 480 (Roche Diagnostics, Basel, Switzerland) and the SYBR Green I detection system (Roche Applied Science, Indianapolis, IN, USA). We normalised gDNA concentrations between individual samples using PCR primers for a distant gene (PTPLA on chromosome 10). Supplementary E, Table 1 lists the key primer pairs used. The EST (DA935254) was present in double the expected copy number in both the patients compared with two healthy unrelated individuals (Figure 4c), consistent with a homozygous duplication of the EST sequence in both patients (ie, they have four copies instead of two). To investigate the size of the putative duplication, we repeated qPCR analysis for genomic regions at variable distances up- and downstream of the EST sequence (Supplementary E, Table 1). These studies indicate that between 40825 and 42937 bp of gDNA, surrounding the EST sequence, is present at twice the normal copy number in gDNA in both patients (Figure 4c). Both parents had 1.5 times the normal copy number, consistent with each having one copy of the chromosome-22 duplication and a normal allele. The break point at the 3′-end is likely located in a 1313 bp region located between regions 3b and 3c (Chr 22:33548218–33549530; GRCh37/hg19 assembly) and the 5′-break point in a 799 bp region located between regions 5b and 5c (Chr 22:33590356–33591154).
Together, these results indicate that the patients are homozygous for a duplication of gDNA around EST (DA935254), and the most likely site of insertion for the duplicated DNA is intron 10 of LARGE, leading to the abnormal splicing of the EST into the LARGE mRNA transcript between exons 10 and 11.
To identify the site of the insertion in intron 10 of LARGE, we designed twelve pairs of overlapping primers (Supplementary E, Figure 2) to cover the whole length of the intron (44117 bp). With two primer pairs (5aF-5aR and 6aF-6aR), we amplified the expected bands in controls and parents but not in the two patients, indicating that the insertion should be in this region (Supplementary E, Figure 2). We repeated the same strategy with several other primer pairs chosen inside this region and in the bordering regions to finally determine a region from 3012 to 4175 bp that could never be amplified in the patients, which likely corresponds to a deleted region associated with the insertion. By qPCR analysis using primer pairs L5F-L6R and L6F-L7R (Supplementary E, Table 1), both parents had reduced amplification compared with controls, consistent with them being heterozygous for a deletion in this region. We were not able to amplify any product with primers chosen on the borders of the insertion and on the non-deleted sides of the intron, and thus did not identify the exact break points for the insertion/deletion rearrangement.
As repetitive sequences are implicated in the pathogenesis of recurrent chromosomal deletion or duplication syndromes (eg, CMT1A),15 we analysed intron 10 and the duplicated region around EST DA935254 for homologous sequences. Both regions shared multiple SINE, LINE and LTR repeats that may have had roles in the genesis of the mutation, but no extended regions of homology were found.
POMT and POMGnT1 activity in patients’ lymphoblast cells
The POMT and POMGnT1 enzymes mediate the first and second steps, respectively, of the O-mannosyl glycosylation of αDG. To investigate whether LARGE mutations may disrupt the glycosylation of αDG by influencing the activity of either POMTs or POMGnT1, we assessed the enzymatic activity of these enzymes in cultured patients’ lymphoblast cells using previously validated assays.16 The POMT activity was based on the amount of [3H]mannose transferred from [3H]mannosylphosphoryldolichol to a glutathione-S-transferase fusion αDG, and the POMGnT1 activity was based on the amount of [3H]GlcNAc transferred from UDP-[3H]GlcNAc to a mannosylpeptide. In brief, we incubated microsomal membrane fractions prepared from patients’ and control EBV-immortalised B lymphocytes, with an acceptor substrate and radiolabelled sugar donor substrate. We then assessed the incorporation of radioactivity into each acceptor substrate. We normalised control and patient samples using the activity level of GnT1, a golgi enzyme involved in the N-glycosylation pathway, which functions independently of O-glycosylation. Results showed no difference between the patient and eight control samples for POMT/GnT1 activity (patient 0.283 vs control 0.287±0.067 SD) or for POMGnT1/GnT1 activity (patient 0.068 vs control 0.071±0.019 SD).
Discussion
In this paper, we report the fourth confirmed family with MDC1D due to mutations in LARGE. The clinical abnormalities are typical for the α-dystroglycanopathy group of disorders. Both sisters have typical features of an α-dystroglycanopathy, such as raised CK levels, dystrophic changes on muscle histology associated with reduced glycosylated α-dystroglycan staining, intellectual disability and brain malformations. In terms of severity of neurological involvement, the two sisters are best classified as having mild MEB disease, even though there were only minor ocular abnormalities. The findings were similar to the first family identified with mutations in LARGE with mild/moderate developmental abnormalities of the cerebellum, pons, brain stem and cerebrum, although our patients are the first ones to be reported with cerebellar cysts.6
Even though standard sequencing of the coding regions of LARGE was normal in the proband of this family, we persisted with analysis of LARGE because there was homozygous linkage to the gene locus in the context of consanguinity. Analysis of cDNA, generated from mRNA derived from patients’ lymphoblast cells, showed an abnormally large product for a PCR reaction that spanned from exons 9 to 12. Sequencing of this product showed that an abnormal sequence of either 197 or 171 bases was spliced between exons 10 and 11 in almost all transcripts. The abnormal sequence is normally situated on chromosome 22 centromeric to the 3′ end of LARGE. In this position, it would not usually be incorporated into the LARGE transcript, even though the sequence contains the necessary splice donor and acceptor sites. To explain its incorporation into the LARGE mRNA, we hypothesised that the EST sequence was translocated into intron 10 of LARGE in our family, possibly through duplication of this genomic region. We assessed the copy number of the EST sequence by qPCR relative to other genes in patients’ gDNA samples and showed twice the expected levels, consistent with a genomic duplication in a homozygous state. Both parents had a 1.5-fold increase in copy number (an increase from two to three copies of the sequence) that was consistent with both of them being asymptomatic heterozygous carriers of the duplication. To map the approximate size of the duplication, we repeated qPCR analysis up- and downstream of the EST sequence and showed the duplication spans between 40 and 43 kb of gDNA. Even though we identified the likely site of insertion in the central part of intron 10 and delimited a large intronic region that cannot be amplified in the patients, mapping the precise break points of the proposed duplication/insertion was not feasible, possibly because of the presence of a more complex rearrangement than anticipated.
Analysis of LARGE mRNA transcripts from patients’ lymphoblast and fibroblast cells suggest that most LARGE mRNA transcripts contain an abnormal sequence. Both versions of the abnormal insertion are predicted to immediately introduce a premature stop codon mid-way through protein translation, just before the second catalytic domain, and result in a non-functional protein (Figure 4b). Small amounts of normally spliced transcript were also seen and the levels appeared slightly higher in fibroblasts than in lymphoblasts, suggesting this may vary in different tissues. Unfortunately, it was not possible to assess levels of normally spliced LARGE mRNA or protein function either in muscle or in brain, the two main tissues that show pathology in the α-dystroglycanopathies. We observed no functionally glycosylated αDG by western blot analysis in patient fibroblast cells, but it is likely that low levels of functional αDG are present in patient tissues at levels below the threshold of detection by western blot for several reasons. Residual glycosylated αDG staining was present in the muscle biopsy, and both sisters have less severe cognitive deficits and brain malformations than a patient with homozygous null mutations in LARGE who had a WWS phenotype.7 We also detected low levels of a normally spliced LARGE gene mRNA transcript in both lymphoblasts and fibroblasts, which should allow normal LARGE protein to be produced at low levels.
We also investigated whether LARGE may exert some of its effects on αDG glycosylation through interactions with either the POMT or the POMGnT1 enzymes. Both POMT and POMGnT1 activities were normal in patients’ lymphoblast cells, strongly suggesting that the two initial steps of αDG glycosylation occur independently of LARGE.
At present, it appears that LARGE is a rare causative of CMD, as the coding regions of the gene have been screened in large cohorts of patients with α-dystroglycanopathy of unknown cause.6, 8 Although experience is very limited at present, it is notable that two of the four confirmed families with CMD, caused by LARGE mutation, have had large gene rearrangements, either a deletion involving several exons7 or a likely intragenic insertion/deletion (our family). In another family, a heterozygous nonsense mutation (p.Trp516X) was identified on standard gene sequencing, leaving open the possibility that a large deletion/duplication or rearrangement on the other allele was overlooked.9 In addition, consanguineous families are described that link to the LARGE locus but no mutation has been found by exon sequencing.7, 17 LARGE was so named for the enormous genomic region it occupies on chromosome 22q12.3 (647 kb) because of the presence of very large introns enriched in low-copy repeats (LCRs). The structure of the LARGE gene may predispose to mutations that involve duplication or deletion of large regions of DNA because of non-allelic homologous recombination between LCRs, the proposed mechanism for many genomic disorders.18 Sequencing of coding regions from gDNA will not identify many large gene rearrangements. Therefore, sequencing LARGE from cDNA and the application of CGH, qPCR, and other methods for detecting rearrangement mutations should be considered routinely.
Accession codes
References
Martin PT : Mechanisms of disease: congenital muscular dystrophies-glycosylation takes center stage. Nat Clin Pract Neurol 2006; 2: 222–230.
Lane PW, Beamer TC, Myers DD : Myodystrophy, a new myopathy on chromosome 8 of the mouse. J Hered 1976; 67: 135–138.
Mathews KD, Rapisarda D, Bailey HL, Murray JC, Schelper RL, Smith R : Phenotypic and pathologic evaluation of the myd mouse. A candidate model for facioscapulohumeral dystrophy. J Neuropathol Exp Neurol 1995; 54: 601–606.
Grewal PK, Holzfeind PJ, Bittner RE, Hewitt JE : Mutant glycosyltransferase and altered glycosylation of alpha-dystroglycan in the myodystrophy mouse. Nat Genet 2001; 28: 151–154.
Lee Y, Kameya S, Cox GA et al: Ocular abnormalities in Large(myd) and Large(vls) mice, spontaneous models for muscle, eye, and brain diseases. Mol Cell Neurosci 2005; 30: 160–172.
Longman C, Brockington M, Torelli S et al: Mutations in the human LARGE gene cause MDC1D, a novel form of congenital muscular dystrophy with severe mental retardation and abnormal glycosylation of alpha-dystroglycan. Hum Mol Genet 2003; 12: 2853–2861.
van Reeuwijk J, Grewal PK, Salih MA et al: Intragenic deletion in the LARGE gene causes Walker-Warburg syndrome. Hum Genet 2007; 121: 685–690.
Mercuri E, Messina S, Bruno C et al: Congenital muscular dystrophies with defective glycosylation of dystroglycan: a population study. Neurology 2009; 72: 1802–1809.
Godfrey C, Clement E, Mein R et al: Refining genotype phenotype correlations in muscular dystrophies with defective glycosylation of dystroglycan. Brain 2007; 130: 2725–2735.
Yoshida-Moriguchi T, Yu L, Stalnaker SH et al: O-mannosyl phosphorylation of alpha-dystroglycan is required for laminin binding. Science 2010; 327: 88–92.
Xiong H, Kobayashi K, Tachikawa M et al: Molecular interaction between fukutin and POMGnT1 in the glycosylation pathway of alpha-dystroglycan. Biochem Biophys Res Commun 2006; 350: 935–941.
Michele DE, Barresi R, Kanagawa M et al: Post-translational disruption of dystroglycan-ligand interactions in congenital muscular dystrophies. Nature 2002; 418: 417–422.
Ervasti JM, Campbell KP : Membrane organization of the dystrophin-glycoprotein complex. Cell 1991; 66: 1121–1131.
Kimura K, Wakamatsu A, Suzuki Y et al: Diversification of transcriptional modulation: large-scale identification and characterization of putative alternative promoters of human genes. Genome Res 2006; 16: 55–65.
Pentao L, Wise CA, Chinault AC, Patel PI, Lupski JR : Charcot-Marie-Tooth type 1A duplication appears to arise from recombination at repeat sequences flanking the 1.5 Mb monomer unit. Nat Genet 1992; 2: 292–300.
Manya H, Bouchet C, Yanagisawa A et al: Protein O-mannosyltransferase activities in lymphoblasts from patients with alpha-dystroglycanopathies. Neuromuscul Disord 2008; 18: 45–51.
Manzini MC, Gleason D, Chang BS et al: Ethnically diverse causes of Walker-Warburg syndrome (WWS): FCMD mutations are a more common cause of WWS outside of the Middle East. Hum Mutat 2008; 29: E231–E241.
Inoue K, Lupski JR : Molecular mechanisms for genomic disorders. Annu Rev Genomics Hum Genet 2002; 3: 199–242.
Acknowledgements
We thank Eric Villard for helpful discussion. NC was supported by a joint grant from the Australian National Health and the Medical Research Council (NHMRC; ID 402861) and the Institut National de la Santé et de la Recherche Médicale. AV and RP by were supported by the Marie Curie grants from the European commission (OIF N°039762 and IIF N°221685). This work was also supported by research grants from the Ministry of Health, Labour and Welfare of Japan (HM, TE; for Nervous and Mental Disorders (20A-13) and Research on Psychiatric and Neurological Diseases and Mental Health), from The Takeda Science Foundation (HM, TE) and from the Paul D Wellstone Muscular Dystrophy Cooperative Research Center grant (1U54NS053672; KPC). KPC is an Investigator of the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on European Journal of Human Genetics website
Rights and permissions
About this article
Cite this article
Clarke, N., Maugenre, S., Vandebrouck, A. et al. Congenital muscular dystrophy type 1D (MDC1D) due to a large intragenic insertion/deletion, involving intron 10 of the LARGE gene. Eur J Hum Genet 19, 452–457 (2011). https://doi.org/10.1038/ejhg.2010.212
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2010.212
Keywords
This article is cited by
-
Muscle satellite cells and impaired late stage regeneration in different murine models for muscular dystrophies
Scientific Reports (2019)
-
Behavioral Responses in Animal Model of Congenital Muscular Dystrophy 1D
Molecular Neurobiology (2016)
-
A review of the diverse genetic disorders in the Lebanese population: highlighting the urgency for community genetic services
Journal of Community Genetics (2015)
-
ISPD loss-of-function mutations disrupt dystroglycan O-mannosylation and cause Walker-Warburg syndrome
Nature Genetics (2012)