Abstract
The 11p15.5 chromosomal region (2.8 Mb) is of particular interest as it encloses five genes (HRAS1, SIRT3, TH, INS and IGF2), the variability of which was found to be associated with life extension by association studies. Mostly important, the above genes are homologous of genes that modulate lifespan in model organisms. We scanned the area in four European sample groups for a total of 1321 centenarians and 1140 younger subjects, who shared with centenarians ethnicity and geographical origin, with a set of 239 SNPs. No significant results (P<0.05) have been found on the earlier associated loci (ie, TH, IGF2, INS and HRAS1), and this study could not confirm the earlier findings on each of those genes. A meta-analysis was carried out on the SIRT3 SNP data; a total number of 2461 samples were included, but no positive association was found except for one SNP having a significant effect (rs939915). The same meta-analysis approach has been applied to the other 229 markers, and six SNPs have been found significant for the frequent genotype (rs4073591, DEAF1-rs4073590, KRTAP5-6-rs11040489, rs4930001, TSPAN32-rs800140 and rs16928120). This experience, although unable to confirm the earlier findings of the literature, highlights all the common difficulties of such studies in human longevity. Despite the rather negative findings presented here, the results derived from unprecedented studies involving such a large number of centenarians should be disseminated, thus contributing to set up adequate strategies to disentangle complex and likely heterogeneous phenotypes.
Similar content being viewed by others
Introduction
In the search for determinants of longevity, we investigated the chromosomal region, 11p.15.5, in four European samples collected from southern Italy (n=92 centenarians, age range 100–108 years old, mean 101.9; and n=97 controls aged 20–60 years old, mean 41.44), northern-central Italy (n=307 centenarians, age range 99–109 years old, mean 101.0; and n=352 controls aged 17–60 years old, mean 41.2), Germany (n=376 centenarians, age range 100–110 years old, mean 104.41; and 376 controls aged 60–75 years old, mean 67.52) and France (n=546 centenarians, age range 99–106 years old, mean 103.1; and 315 controls aged 18–70 years old, mean 51.2). We analyzed a total of 1321 centenarians (232 males and 1089 females) and 1140 younger subjects (530 males and 610 females), who shared ethnicity and geographical origin with the centenarians.
The 11p15.5 area is of particular interest as it encloses five genes (HRAS1, SIRT3, TH, INS and IGF2), the variability of which was found to be associated with human longevity.1, 2, 3, 4, 5, 6, 7 Most importantly, the above genes are homologous of genes that are known to modulate lifespan in model organisms (http://genomics.senescence.info/). As for HRAS1, an earlier study of a central-northern Italian sample showed that the frequency of a particular VNTR allele decreases in centenarians compared with that in young people, and it was estimated that, during ageing, carriers of this allele have a relative mortality risk of 1.126, with a 95% CI of 1.044–1.213.2 Likewise, by analyzing the genotype-specific survival function relevant to the G477T marker of SIRT3 in a southern Italian population, we found a male-specific association between the marker and survival in the elderly.7 A subsequent study showed that the association was because of linkage disequilibrium between the G477T marker and a VNTR allele-specific enhancer located in intron 5 of SIRT3. In particular, the 2b allele was absent in male centenarians, whereas it was significantly more frequent in younger males.1 Similarly, in both southern and northern Italian males, the functional microsatellite polymorphism (TCAT)n located in intron A of the TH gene showed association with life extension.4, 8 In addition, the entire TH-INS-IGF2 region was found to be associated with longevity in southern Italians; in particular, the frequency distributions of TH-INS haplotypes and of TH-IGF2 haplotypes were significantly different between centenarians and younger controls in both females and males.5
On the whole, the above findings indicated that, at least in the investigated populations, and in some cases in a sex-specific manner, the 11p15.5 region harbors gene variants associated with life extension.
Therefore, instead of focusing on candidate genes, the aim of this study was to extend the earlier analyzes by investigating the entire 11p15.5 region (2.4 Mb) with a set of suitable single-nucleotide polymorphisms (SNPs).
Materials and methods
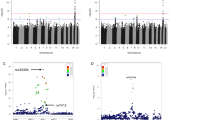
A set of 239 SNPs (tagSNPs and htSNPs) were selected on the basis of HapMap data from CEPH trios that were found to be suitable for analysis using SNPlex technology (Applied Biosystems, Foster City, CA, USA). The obtained genotypes were tested for Hardy–Weinberg equilibrium and the three populations (Central Italy, South Italy and Germany) were subjected to AMOVA,9 which suggested not to pool the three population data. The genotyping results were analyzed using three different approaches: single-point analysis, sliding window multi-point regression analysis corrected with permutations10 and permutation analysis of the haplotype statistics.11
Results
A number of positive associations were observed and filtered through the different analysis approaches, to correct for multiple testing and to take into account the different powers of the analysis methods.12 However, the results were not concordant in the three populations (Central Italy, South Italy and Germany).
No significant results (P<0.05) were found for any of the earlier associated loci (ie TH, IGF2, INS and HRAS1).1, 2, 3, 4, 6, 7, 8, 13, 14, 15, 16, 17, 18
An exception was SIRT3, for which different markers showed significant association signals in the populations, that is, the association was present in Italian females and German males.
Among the different genes, a specific interest was thus focused on the SIRT3 locus, given the extensive literature, suggesting an association of sirtuin genes with longevity19 and mitochondrial functionality,16 an important topic in ageing and longevity.20
A replication study was then designed for SIRT3: eight SNPs were tested in a French sample (546 cases–age range 99–106 years old, mean 103.1, and 315 controls–age range 18–70 years old, mean 51.2) at CEPH in Paris. None of the earlier findings were confirmed in this analysis.
To increase the power of the analysis and to gain a deeper insight into the genotype data, a meta-analysis was carried out on the SIRT3 SNP data. A total number of 2461 samples were included by using the inverse variance weighting for pooling in the presence of a fixed-effects model,21, 22 and the Der Simonian–Laird estimation in the random-effects model.23 The analysis was carried out at the level of the rare and frequent genotypes, and at the heterozygous and allelic level. No positive association was found except for one SNP having a significant effect (rs939915); unfortunately, the genotyping of this marker in central Italians resulted in a considerable number of dropouts and the false discovery rate (q-value of 0.145) indicates that this result is not robust enough (Table 1).
The same meta-analysis approach was also applied to the other markers for a better understanding of the Italian and German population results. A meta-analysis of all the 239 11p15.5 markers on 1600 samples was then carried out. The results were filtered by applying a false discovery rate analysis24, 25, 26 to the P-values resulting from the model adopted for each marker in the meta-analysis.
Six SNPs were found to be significant for the frequent genotypes (rs4073591, DEAF1-rs4073590, KRTAP5-6-rs11040489, rs4930001, TSPAN32-rs800140 and rs16928120) at a q-value ⩽0.36, confirming a few of the positive associations obtained in the original first three analysis approaches. Data on KRTAP5-6_rs11040489, rs4930001, TSPAN32_rs800140 and rs16928120 were significant in the allelic analysis too (Table 2).
Two findings were considered interesting: DEAF1 encodes a transcription factor binding the retinoic acid response element and activating the proenkephalin gene; when secreted, it is a suppressin, and stops cells in G0/G1. TSPAN32 codes for a tumor-suppressing fragment, and is located in the imprinted region of 11p15.5.
Despite the biological interest of these results, each of the positive findings requires a follow-up analysis, and no conclusion can be made at present.
Discussion
This work represents the results of the analysis on an unprecedented collection of centenarians and controls, and it can be considered the most extensive investigation ever carried out in a case–control study in a specific candidate chromosomal region in ageing.
This experience, although unable to confirm the earlier findings, highlights all the common difficulties of such studies in human longevity.
The methodological (samples from different populations) and statistical (multiple analysis correction, false-positive rate, meta-analysis criteria, among others) problems that become apparent when a complex trait such as longevity is addressed represent a hot topic in this field. Our choice was focused on combining different methods rather than discarding useful information. Instead of compromising on power, we filtered the signals through the consistency of the different analyses and by applying a false discovery rate approach to the final results.
The differences between the South and Central Italian and the German populations highlight the importance of taking into account population heterogeneity in this kind of studies.
Those studies on ageing, which take into consideration demographic data, suggest that different susceptibility genes are likely to underlie the longevity phenotype in genetically different populations. The consequences are crucial with regard to the results (and interpretation) of replication studies.27 Our study is not the only one highlighting contrasting results from different populations, and we suggested that centenarians might be considered phenocopies who display the same phenotype (longevity), but who have attained longevity by taking advantage of different gene/environment contributions.27 In fact, longevity, more than any other complex trait, may result from a lifelong (more than 100 years) interaction between genetic and environmental factors. Moreover, epigenetics and stochasticity could also contribute to longevity, but such topics are still largely unexplored in humans.
As the number of males among centenarians is very low,28 it is difficult to set up gender-matched samples. The choice is then to have samples representative of the overall. Although earlier findings clearly indicate gender-specific associations, a stratification by gender in these centenarian populations leads to unacceptable power in the analyzes.
Moreover, as the results of the association studies in the German, Central Italian and Southern Italian populations were different, we thought it worthwhile to carry out additional genotyping in the French cohort to reach more clear-cut results. However, the results deflate the significance, reinforcing the interpretation that we are likely to deal with population specificity.
The case–control study design has limitations in highlighting polymorphisms displaying U-shape patterns of genotype frequencies. The groups of Franceschi and De Benedictis27, 29, 30, 31, 32 earlier stressed the likelihood of a genetic variant to exert different effects at different ages, and a cross-sectional study design in which two age groups are compared may not be able to identify all variants potentially associated with such a complex phenotype.
Despite the rather negative findings presented here, we are of the opinion that results deriving from unprecedented studies involving such a large number of centenarians should be disseminated, thus contributing to set up adequate strategies to disentangle complex and likely heterogeneous phenotypes.
References
Bellizzi D, Rose G, Cavalcante P et al: A novel VNTR enhancer within the SIRT3 gene, a human homologue of SIR2, is associated with survival at oldest ages. Genomics 2005; 85: 258–263.
Bonafe M, Barbi C, Olivieri F et al: An allele of HRAS1 3′variable number of tandem repeats is a frailty allele: implication for an evolutionarily-conserved pathway involved in longevity. Gene 2002; 286: 121–126.
Bonafe M, Barbieri M, Marchegiani F et al: Polymorphic variants of insulin-like growth factor I (IGF-I) receptor and phosphoinositide 3-kinase genes affect IGF-I plasma levels and human longevity: cues for an evolutionarily conserved mechanism of life span control. J Clin Endocrinol Metab 2003; 88: 3299–3304.
De Benedictis G, Carotenuto L, Carrieri G et al: Gene/longevity association studies at four autosomal loci (REN, THO, PARP, SOD2). Eur J Hum Genet 1998; 6: 534–541.
De Luca M, Rose G, Bonafe M et al: Sex-specific longevity associations defined by tyrosine hydroxylase-insulin-insulin growth factor 2 haplotypes on the 11p15.5 chromosomal region. Exp Gerontol 2001; 36: 1663–1671.
Gaunt TR, Cooper JA, Miller GJ, Day IN, O'Dell SD : Positive associations between single nucleotide polymorphisms in the IGF2 gene region and body mass index in adult males. Hum Mol Genet 2001; 10: 1491–1501.
Rose G, Dato S, Altomare K et al: Variability of the SIRT3 gene, human silent information regulator Sir2 homologue, and survivorship in the elderly. Exp Gerontol 2003; 38: 1065–1070.
Tan Q, Bellizzi D, Rose G et al: The influences on human longevity by HUMTHO1.STR polymorphism (tyrosine hydroxylase gene). A relative risk approach. Mech Ageing Dev 2002; 123: 1403–1410.
Excoffier L, Laval LG, Scheider S : Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol Bioinform Online 2005; 1: 47–50.
Purcell S, Daly MJ, Sham PC : WHAP: haplotype-based association analysis. Bioinformatics 2007; 23: 255–256.
Barrett JC, Fry B, Maller J, Daly MJ : Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005; 21: 263–265.
Becker T, Cichon S, Jonson E, Knapp M : Multiple testing in the context of haplotype analysis revisited: application to case–control data. Ann Hum Genet 2005; 69: 747–756.
Barbieri M, Bonafe M, Franceschi C, Paolisso G : Insulin/IGF-I-signaling pathway: an evolutionarily conserved mechanism of longevity from yeast to humans. Am J Physiol Endocrinol Metab 2003; 285: E1064–E1071.
Barbieri M, Ferrucci L, Ragno E et al: Chronic inflammation and the effect of IGF-I on muscle strength and power in older persons. Am J Physiol Endocrinol Metab 2003; 284: E481–E487.
Franceschi C, Olivieri F, Marchegiani F et al: Genes involved in immune response/inflammation, IGF1/insulin pathway and response to oxidative stress play a major role in the genetics of human longevity: the lesson of centenarians. Mech Ageing Dev 2005; 126: 351–361.
Onyango P, Celic I, McCaffery JM, Boeke JD, Feinberg AP : SIRT3, a human SIR2 homologue, is an NAD-dependent deacetylase localized to mitochondria. Proc Natl Acad Sci USA 2002; 99: 13653–13658.
Tanaka Y, Minami Y, Mine S et al: H-Ras signals to cytoskeletal machinery in induction of integrin-mediated adhesion of T cells. J Immunol 1999; 163: 6209–6216.
Vega A, Salas A, Costas J, Barros F, Carracedo A : Length variability and interspersion patterns of the HRAS1 minisatellite: a new approach for the reconstruction of human population relationships. Ann Hum Genet 2001; 65: 351–361.
Guarente L, Picard F : Calorie restriction – the SIR2 connection. Cell 2005; 120: 473–482.
Salvioli S, Capri M, Santoro A et al: The impact of mitochondrial DNA on human lifespan: a view from studies on centenarians. Biotechnol J 2008; 3: 740–749.
Fleiss JL : The Mantel–Haenszel estimator in case–control studies with varying numbers of controls matched to each case. Am J Epidemiol 1984; 120: 1–3.
Mantel N, Haenszel W : Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 1959; 22: 719–748.
DerSimonian R, Laird N : Meta-analysis in clinical trials. Control Clin Trials 1986; 7: 177–188.
Storey J, Taylor J, Siegmund D : Strong control, conservative point estimation, and simultaneous conservative consistency of false discovery rates: A unified approach. J R Stat Soc Series B 2004; 66: 187–205.
Storey JD : A direct approach to false discovery rates. J R Stat Soc Series B 2002; 64: 479–498.
Storey JD, Tibshirani R : Statistical significance for genome wide studies. Proc Natl Acad Sci USA 2003; 100: 9440–9445.
De Benedictis G, Franceschi C : The unusual genetics of human longevity. Sci Aging Knowledge Environ 2006; 2006: 20.
Passarino G, Calignano C, Vallone A et al: Male/female ratio in centenarians: a possible role played by population genetic structure. Exp Gerontol 2002; 37: 1283–1289.
Cardelli M, Cavallone L, Marchegiani F et al: A genetic-demographic approach reveals male-specific association between survival and tumor necrosis factor (A/G)-308 polymorphism. J Gerontol A Biol Sci Med Sci 2008; 63: 454–460.
Dato S, Carotenuto L, De Benedictis G : Genes and longevity: a genetic-demographic approach reveals sex- and age-specific gene effects not shown by the case-control approach (APOE and HSP70.1 loci). Biogerontology 2007; 8: 31–41.
Yashin AI, De Benedictis G, Vaupel JW et al: Genes, demography, and life span: the contribution of demographic data in genetic studies on aging and longevity. Am J Hum Genet 1999; 65: 1178–1193.
Yashin AI, De Benedictis G, Vaupel JW et al: Genes and longevity: lessons from studies of centenarians. J Gerontol A Biol Sci Med Sci 2000; 55: B319–B328.
Acknowledgements
We thank the GEHA – Genetics of Healthy Ageing Consortium, supported under the Sixth Framework Programme for RTD of the European Commission – Priority 1. Life sciences, Genomics and Biotechnology for health (contract no. LSHM-CT-2004-503270).
For excellent technical assistance, we acknowledge the laboratory personnel of the Institute of Clinical Molecular Biology at the Christian-Albrechts-University Kiel.We thank the Biological Ressource Center at the Fondation Jean Dausset-CEPH for preparing and genotyping DNA samples of the French Population.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lescai, F., Blanché, H., Nebel, A. et al. Human longevity and 11p15.5: a study in 1321 centenarians. Eur J Hum Genet 17, 1515–1519 (2009). https://doi.org/10.1038/ejhg.2009.54
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2009.54
Keywords
This article is cited by
-
The CEPH aging cohort and biobank: a valuable collection of biological samples from exceptionally long-lived French individuals and their offspring for longevity studies
GeroScience (2023)
-
Sirtuins: Role in the Regulation of Oxidative Stress and the Pathogenesis of Neurodegenerative Diseases
Neuroscience and Behavioral Physiology (2022)
-
The dominant model analysis of Sirt3 genetic variants is associated with susceptibility to tuberculosis in a Chinese Han population
Molecular Genetics and Genomics (2020)
-
Sirtuin 3 deficiency does not alter host defenses against bacterial and fungal infections
Scientific Reports (2017)
-
Insulin/insulin-like growth factor-1 signalling (IIS) based regulation of lifespan across species
Biogerontology (2017)