Abstract
In their studies on the molecular basis of osteopoikilosis, Menten et al have identified three individuals with microdeletions on chromosome 12q14.4, which removed several genes including LEMD3, the osteopoikilosis gene. In addition to osteopoikilosis, affected individuals had growth retardation and developmental delay. We now report a smaller 12q14.4 microdeletion in a boy with severe pre and postnatal growth failure, and mild developmental delay; the patient was small at birth and presented with poor feeding and failure to thrive during the first 2 years of life, similar to the phenotype of primordial dwarfism or severe Silver-Russell syndrome (SRS). The 12q14 deletion did not include LEMD3, and no signs of osteopoikilosis were observed on skeletal radiographs. Among the deleted genes, HMGA2 is of particular interest in relationship to the aberrant somatic growth in our patient, as HMGA2 variants have been linked to stature variations in the general population and loss of function of Hmga2 in the mouse results in the pygmy phenotype that combines pre and postnatal growth failure, with resistance to the adipogenic effect of overfeeding. Sequencing of the remaining HMGA2 allele in our patient showed a normal sequence, suggesting that HMGA2 haploinsufficiency may be sufficient to produce the aberrant growth phenotype. We conclude that the 12q14.4 microdeletion syndrome can occur with or without deletion of LEMD3 gene; in LEMD3-intact cases, the phenotype includes primordial short stature and failure to thrive with moderate developmental delay, but osteopoikilosis is absent. Such cases will likely be diagnosed as Silver-Russell-like or as primordial dwarfism.
Similar content being viewed by others
Introduction
Osteopoikilosis denotes the presence of dense osseous lesions throughout the skeleton. These lesions are easily identified on skeletal radiographs of the limbs as small hyperdense spots. Although osteopoikilosis usually remains without any clinical consequence, in some individuals more significant sclerotic bone lesions develop that are called melorheostosis. When osteopoikilosis or melorheostosis is associated with skin changes, the phenotype is called Buschke-Ollendorff syndrome. Familial cases of these three conditions are often caused by dominant mutations in the LEMD3 gene,1 whereas the molecular basis of sporadic cases often remains undetermined.
In the course of studying the molecular basis of osteopoikilosis, Menten et al2 have observed three patients who had a more complex phenotype including short stature, failure to thrive, and moderate mental retardation. These three individuals were found to have deletions involving the 12q14.4 region that included LEMD3, as well as several other genes. Menten et al2 concluded that these patients proved the existence of a ‘12q14 microdeletion syndrome’.
Although osteopoikilosis could be attributed to LEMD3 haploinsufficiency, the other clinical components, that is, primordial short stature and developmental delay could not be attributed with certainty since none of the 20 or so other genes present in the common deleted region were known disease genes in the human, although a possible role of the HMGA2 gene in growth failure was noted. Here, we report on a patient with a microdeletion of 12q14 ascertained because of pre and postnatal growth failure; he did not have osteopoikilosis and his deletion, though overlapping the three earlier reported ones, was significantly smaller and did not encompass the LEMD3 gene, but did encompass the HMGA2 gene, thus further supporting the role of haploinsufficiency of HMGA2 in the etiology of short stature and failure to thrive.
Materials and methods
After informed consent was obtained, blood samples were drawn from the proband and from his parents and genomic DNA was extracted from peripheral leucocytes using the Gentra genomic DNA blood isolation kit (Gentra, Minneapolis, MN, USA) according to the manufacturer's protocol.
Affimetrix SNP-array
The GeneChip® Human Mapping 50 K Array Xba 240 (Affymetrix, Santa Clara, CA, USA) was utilized. The hybridization was carried out according to the manufacturer's protocol. The chips were washed with the Fluidics Station 450 and the probe arrays were scanned using the GeneChip® Scanner 3000 7 G according to the manufacturer's recommendations (Affymetrix). The data were analyzed with the program CNAT (Affymetrix) to look for LOH in the patient compared with his parents as well as 100 control samples (data provided by Affymetrix).
Agilent array-CGH 244K
The aCGH has been carried out using the Agilent platform according to the manufacturer's instructions. The data were analyzed utilizing the CGH Analytics Software (Agilent Technologies, Inc., Santa Clara, CA, USA) and the Human May 2004 (hg17) Assembly.
HMGA2 sequence analysis
Genomic DNA of 100 ng was used as template in the PCR reaction. Primers were designed to amplify all 5 coding exons and the 5′- and 3′-flanking regions of the more abundant form, and the alternative exons of the additional 5 splice-variants (isoforms b-f, exons a-e). The PCR reaction was carried out with the PCR Master Mix (Promega, Madison, WI, USA) in a 25 μl reaction at an annealing temperature of 58°C for most exons (see Supplementary Table 1 for primer sequences and annealing temperature). In addition the PCR reactions of exon 1 were supplemented with 5% glycerol and PCR reactions of exons 3e and 5 with 5 μl of betaine 5M. The PCR products were purified using the Witzard PCR purification Kit (Promega) and directly sequenced with Dye Terminator v3.1 on a 3130 Genetic Analyzer (Applied Biosystems, Forster City, CA, USA) according to the manufacturer's recommendations.
Results
Clinical description
The boy was the third child of apparently healthy parents of Rumanian origin who were first-degree cousins. One brother was healthy, whereas one sister had died with congenital heart disease. The parents were 33 yrs old at the time of delivery; their heights were 155 cm (father) and 160 cm (mother). The proband was delivered by caesarean section at week 36 because of the observation of severe intrauterine growth retardation that was, at that time, tentatively attributed to maternal glucose intolerance and hypertension. Birth weight was 1730 gr (<10th percentile), length 43 cm (−4 SD), OFC 29 cm (−3.66 SD). Apgar scores were 7 and 9 at 1 and 5 min, respectively. The baby showed moderate respiratory distress and feeding difficulties and was fed by nasogastric tube. There were transient hypoglycaemia and hypocalcaemia. An echocardiogram showed patent ductus arteriosus which closed at 2 months of age after pharmacological therapy. At the age of 2.5 months he was again hospitalized because of recurrent respiratory infections and the presence of cyanotic and hypertonic crises. At 10.5 months he was alert but could not yet sit unsupported. An echocardiogram at 12 months of age showed a ventricular septal defect.
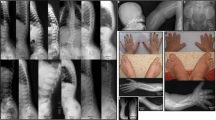
At age 18 months, he was referred for severe proportionate short stature (Figure 1a): his length was 65 cm (−5.3 SD), weight 5070 gr (−4.9 SD) and OFC 42 cm (−5.6 SD). He had a very poor appetite and was still fed by nasogastric tube. He showed generalized muscular hypotonia with good eye contact. He was able to sit unsupported but not able to walk. He had a triangular face with prominent forehead, down-turned corners of his mouth, a high-vaulted palate, and slight micrognathia (Figure 1) and a clinical diagnosis of Silver-Russell was considered at that time; there was no body asymmetry. He was a very small but good-humored boy; the parents' main worry was his lack of appetite that they felt was responsible for the poor growth and developmental delay.
The two photographs show the clinical phenotype of the child with 12q14 microdeletion. Note short stature, reduced adipose tissue, muscular hypotrophy, and small, triangular face. The severe pre- and postnatal growth delay is illustrated by the curves for height (left) and head circumference (OFC); it is interesting that, growth velocity is normal, but there is no catch-up growth. Weight was also severely reduced (see text). The two radiographs (taken at age 2 yrs 4 months) show moderate osteopenia, slight coarsening of the bone trabeculae, and markedly delayed skeletal maturation (bone age of approximately 6 months) but absence of osteopoikilosis.
Karyotype analysis at that time showed the presence of a de novo rearrangement characterized by the insertion in 1q43 of bands 4p11–p15.31 (data not shown). At age 3 yrs, his height was 77 cm (−4.85 SD), his weight 6500 gr (−5 SD) and his OFC 44.5 cm (−5.1 SD) (Figure 1b) He was still not completely able to walk unaided; he showed some attention deficit with hyperactivity and he was not speaking but only babbling. At age 3 yrs 7 months, an arginine stimulation test showed a suboptimal growth hormone response, and the patient was started on growth hormone treatment; after 7 months of treatment, the growth velocity did not change but his overall conditions seemed to improve slightly.
Molecular data
Given the presence of a chromosomal aberration in the proband, we wanted to exclude imbalances at the breakpoints that could explain the phenotype. We thus carried out an Affymetrix SNP array with an average spatial resolution of 50 Kb. Against our expectations, the study did not identify any genomic imbalance around the insertion breakpoints but instead showed an interstitial deletion of about 1.83 Mb on chromosome 12 (data not shown).
To confirm the array data and in order to define the breakpoints, array-CGH experiments using an oligonucleotides array with an averaged spatial resolution of approximately 10–12 kb were carried out in the patient and his parents (Agilent 244K). The presence of the 1.8 Mb 12q14.3 deletion in the proband was confirmed (Figure 2), whereas both parents showed a normal result. The proximal breakpoint mapped to 12q14.3, with the last present oligonucleotide located in 64.46 Mb and the first deleted in 64.47 Mb. The distal breakpoint is located in 12q15 between 66.27 and 66.31 Mb (last oligonucleotide deleted and first present, respectively). The deletion comprises 6 known genes (HELB, TMBIM4, IRAK3, CAND1, GRIP1, and HMGA2) (Figure 3).
The deletion indicated by the array CGH experiment is mapped against the corresponding genomic region in the UCSC genome browser. In the lower part, the extent of the deletion in the patient described in this report is compared with that in the 3 patients reported by Menten et al (2007); note preservation of the osteopoikilosis gene LEMD3 in the present patient.
HMGA2 mutation analysis in the proband did not show any sequence changes in the remaining allele. The proband was hemizygous for the T allele at the SNP rs1042725, located in the 3′-UTR region of the gene and associated with height variability in the general population.3
Discussion
Menten et al recently delineated a novel 12q14 microdeletion syndrome in three unrelated patients who were ascertained for osteopoikilosis but who also had pre- and postnatal short stature and developmental delay.2 The facial features of our patient resembles those observed in Patient 1 in the paper by Menten et al.2 The 12q14.4 deletions in those three patients ranged from 3.44 to 6 Mb. A similar combination of findings (osteopoikilosis, short stature and developmental delay) had been reported in an adult man by Jurenka and van Allen in 1995;4 although DNA from that patient was not available for study,2 it might be possible that that patient also had a 12q14 microdeletion.
We now report on a boy ascertained for prenatal short stature and failure to thrive who has a 12q14 microdeletion that overlaps with those of the patients reported by Menten et al,2 but is only approximately 1.8 Mb and does not include the LEMD3 gene; the clinical phenotype is similar with the exception of osteopoikilosis that is absent in our patient. Thus, while osteopoikilosis is a useful diagnostic sign when present, the cardinal features of the microdeletion syndrome seem to be pre- and postnatal growth failure and mild to moderate developmental delay with microcephaly (Table 1)
The 12q14 deletion in our patient removes 6 genes: HELB, TMBIM4, IRAK3, CAND1, GRIP1, and HMGA2 (Figure 2). Information on some of these genes is scanty, only the latter 4 genes are listed in OMIM, and genotype-phenotype correlations are still speculative. HELB codes for a helicase (DNA) B that seems to be required for S-phase entry. TMBIM4 stands for transmembrane BAX inhibitor motif containing protein 4; it is also known as S1R, and its function is unknown. CAND1 or TBP-interacting protein seems to be a ubiquitine ligase complex regulator6. GRIP1 encodes a protein that contains a PDZ domain, important for synaptic function,7 and that is highly expressed in both fetal and adult brain. In theory, haploinsufficiency could account for the neurodevelopmental aspect of the microdeletion syndrome. For these 4 genes, no genotype–phenotype associations are known so far. IRAK3 codes for interleukin 1 receptor-associated kinase M, which is involved in host defence and has been associated with asthma susceptibility.8
More information is available on the HGMA2 gene, and indeed, as already noted by Menten et al,2 that gene is an interesting candidate to explain the aberrant growth pattern seen in the 12q14 microdeletion syndrome. Earlier known as HMGIC, HMGA2 encodes a protein that belongs to the non-histone chromosomal high-mobility group (HMG) protein family. HMGA2 contains 3 AT-hook motifs, involved in DNA binding, encoded by the first 3 exons; a 11-amino acid sequence characteristic of HMGA2 and absent in the other family proteins (linker domain) encoded by exon 4 and part of exon 5; and an acidic C-terminal domain encoded by the fifth exon.9 One principal full-length transcript as well as 5 additional splice-variants are known (isoforms b-f); the latter isoforms lack exons 4 and 5 which are replaced by sequences derived from parts of intron 3.10, 11
Both gain and loss of function variants of HMGA2 have been observed in biological systems. Translocations involving HMGA2 are commonly found in mesenchymal tumors, especially lipomas.12 The chromosomal breakpoint frequently lies within the large intron 3, and thus, the rearrangement determines the formation of a chimeric protein containing the AT hooks of HMGA2 attached to ‘illegitimate’ carboxy terminals. Transgenic mice carrying a truncated HMGA2 protein, deprived of its acidic tail, develop an overgrowth phenotype with abdominal and pelvic lipomatosis, suggesting a selective advantage for removal of the acidic carboxy terminal in rapidly proliferating neoplastic cells.13 The observation of high expression of a HMGA2 variant lacking the linker region and the acidic carboxy terminal domain in mesenchymal tumors supports this hypothesis.14 A patient with overgrowth and lipomas has been reported to have a de novo chromosomal inversion with one breakpoint lying within the intron 3 of the HMGA2 gene.15
Loss of function of HMGA2, on the other hand, has been linked to dwarfism in the mouse. A long-known spontaneous mouse mutant with reduced growth, pygmy, has been linked to a series of different loss of function mutations around the Hmga2 locus, and insertional mutagenesis at the same locus reproduces the pygmy phenotype.16 Notably, the pygmy trait is semidominant, with heterozygotes exhibiting a milder growth reduction than homozygotes. More recently, the importance of HMGA2 as a growth regulator in the human has come from a genome-wide association study for stature variation in healthy children and adults.3, 17 These studies have shown that a common SNP in HMGA2 is reproducibly associated with height variation in the general population. In addition to reduced body size, Hmga2 mutant mice show a disproportionate reduction in body weight.16 This observation, together with the lipomatosis phenotype observed in the gain-of-function state (see above), suggests that HMGA2 plays a role not only in growth and development but also in adipogenesis. In support of this hypothesis is the observation of Anand et al,18 who found that Hmga2 knock-out mice are resistant to diet-induced obesity and that reduction of Hmga2 expression protects mice from leptin-deficiency-induced obesity. Thus, there is circumstantial but strong evidence suggesting that HMGA2 haploinsufficiency may be responsible both for reduced growth seen in the 12q14 microdeletion patients and for the poor appetite, failure to thrive and lack of adipose tissue as illustrated by our patient.
Finally, it must be noted cautiously that growth in our patient may also be influenced by the short stature of his father; and that we cannot rule out a possible phenotypic contribution of the de novo insertion in 1q43 of bands 4p11-p15.31, although the array-CGH did not show any imbalance at the breakpoints.
In conclusion, the observation of a smaller 12q14.4 deletion that recapitulates the phenotype described by Menten et al2 with the exception of osteopoikilosis refines the definition of the 12q14.4 microdeletion syndrome and reinforces the notion of a major effect of HMGA2 on pre- and postnatal growth in the human. More observations are needed to elucidate the role of individual genes in the pathogenesis of growth failure as well as of developmental delay. Four of the 5 patients with the microdeletion 12q14 (including the patient reported by Jurenka and Van Allen)2, 4 had the rare sign of osteopoikilosis; patients without that sign are likely to be diagnosed as less specific conditions such as primordial dwarfism or the Silver–Russell syndrome; such patients are not uncommon, and the incidence of the 12q14 microdeletion syndrome could be more frequent than indicated by the osteopoikilosis-positive patients only. The 12q14 microdeletion findings also suggest that HMGA2 is a candidate gene for conditions featuring pre- and postnatal growth failure and failure to thrive that is otherwise unexplained.
References
Hellemans J, Preobrazhenska O, Willaert A et al: Loss-of-function mutations in LEMD3 result in osteopoikilosis, Buschke-Ollendorff syndrome and melorheostosis. Nat Genet 2004; 36: 1213–1218.
Menten B, Buysse K, Zahir F et al: Osteopoikilosis, short stature and mental retardation as key features of a new microdeletion syndrome on 12q14. J Med Genet 2007; 44: 264–268.
Weedon MN, Lettre G, Freathy RM et al: A common variant of HMGA2 is associated with adult and childhood height in the general population. Nat Genet 2007; 39: 1245–1250.
Jurenka SB, Van Allen MI : Mixed sclerosing bone dysplasia, small stature, seizure disorder, and mental retardation: a syndrome? Am J Med Genet 1995; 57: 6–9.
Friedman JM, Baross A, Delaney AD et al: Oligonucleotide microarray analysis of genomic imbalance in children with mental retardation. Am J Hum Genet 2006; 79: 500–513.
Liu J, Zhang T, Lu T et al: DNA-binding and cleavage studies of macrocyclic copper(II) complexes. J Inorg Biochem 2002; 91: 269–276.
Hata Y, Nakanishi H, Takai Y : Synaptic PDZ domain-containing proteins. Neurosci Res 1998; 32: 1–7.
Balaci L, Spada MC, Olla N et al: IRAK-M is involved in the pathogenesis of early-onset persistent asthma. Am J Hum Genet 2007; 80: 1103–1114.
Chau KY, Patel UA, Lee KL, Lam HY, Crane-Robinson C : The gene for the human architectural transcription factor HMGI-C consists of five exons each coding for a distinct functional element. Nucleic Acids Res 1995; 23: 4262–4266.
Hauke S, Flohr AM, Rogalla P, Bullerdiek J : Sequencing of intron 3 of HMGA2 uncovers the existence of a novel exon. Genes Chromosomes Cancer 2002; 34: 17–23.
Hauke S, Leopold S, Schlueter C et al: Extensive expression studies revealed a complex alternative splicing pattern of the HMGA2 gene. Biochim Biophys Acta 2005; 1729: 24–31.
Schoenmakers EF, Wanschura S, Mols R, Bullerdiek J, Van den Berghe H, Van de Ven WJ : Recurrent rearrangements in the high mobility group protein gene, HMGI-C, in benign mesenchymal tumours. Nat Genet 1995; 10: 436–444.
Battista S, Fidanza V, Fedele M et al: The expression of a truncated HMGI-C gene induces gigantism associated with lipomatosis. Cancer Res 1999; 59: 4793–4797.
Ashar HR, Tkachenko A, Shah P, Chada K : HMGA2 is expressed in an allele-specific manner in human lipomas. Cancer Genet Cytogenet 2003; 143: 160–168.
Ligon AH, Moore SD, Parisi MA et al: Constitutional rearrangement of the architectural factor HMGA2: a novel human phenotype including overgrowth and lipomas. Am J Hum Genet 2005; 76: 340–348.
Zhou X, Benson KF, Ashar HR, Chada K : Mutation responsible for the mouse pygmy phenotype in the developmentally regulated factor HMGI-C. Nature 1995; 376: 771–774.
Weedon MN, Lango H, Lindgren CM et al: Genome-wide association analysis identifies 20 loci that influence adult height. Nat Genet 2008; 40: 575–583.
Anand A, Chada K : In vivo modulation of Hmgic reduces obesity. Nat Genet 2000; 24: 377–380.
Acknowledgements
This study was supported by the German BMBF Rare Diseases network SKELNET (project GFGN01141901) and by the European Community (FP6, ‘EuroGrow’ project, LSHM-CT-2007–037471). Dr. Mari was recipient of a study grant from the Deutscher Akademischer Austauschdienst (DAAD) and of a research grant from the University of Siena (PAR 2006).
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary information
Rights and permissions
About this article
Cite this article
Mari, F., Hermanns, P., Giovannucci-Uzielli, M. et al. Refinement of the 12q14 microdeletion syndrome: primordial dwarfism and developmental delay with or without osteopoikilosis. Eur J Hum Genet 17, 1141–1147 (2009). https://doi.org/10.1038/ejhg.2009.27
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2009.27
Keywords
This article is cited by
-
12q14.3 microdeletion involving HMGA2 gene cause a Silver-Russell syndrome-like phenotype: a case report and review of the literature
Italian Journal of Pediatrics (2020)
-
Die Rolle der HMGA-Proteine („high mobility group AT-hook“) bei Proliferation und Differenzierung mesenchymaler Zellen und Gewebe
Gefässchirurgie (2020)
-
Variations in the high-mobility group-A2 gene (HMGA2) are associated with idiopathic short stature
Pediatric Research (2016)
-
Identifying candidate genes for discrimination of ulcerative colitis and Crohn’s disease
Molecular Biology Reports (2014)
-
The Lin28b–let-7–Hmga2 axis determines the higher self-renewal potential of fetal haematopoietic stem cells
Nature Cell Biology (2013)