Abstract
Non-dystrophic myotonias (NDMs) are caused by mutations in CLCN1 or SCN4A. The purpose of the present study was to optimize the genetic characterization of NDM in The Netherlands by analysing CLCN1 and SCN4A in tandem. All Dutch consultant neurologists and the Dutch Patient Association for Neuromuscular Diseases (Vereniging Spierziekten Nederland) were requested to refer patients with an initial diagnosis of NDM for clinical assessment and subsequent genetic analysis over a full year. Based on clinical criteria, sequencing of either CLCN1 or SCN4A was performed. When previously described mutations or novel mutations were identified in the first gene under study, the second gene was not sequenced. If no mutations were detected in the first gene, the second gene was subsequently also analysed. Underlying NDM mutations were explored in 54 families. In total, 20% (8 of 40) of our probands with suspected chloride channel myotonia showed no CLCN1 mutations but subsequent SCN4A screening revealed mutations in all of them. All 14 probands in whom SCN4A was primarily sequenced showed a mutation. In total, CLCN1 mutations were identified in 32 families (59%) and SCN4A in 22 (41%), resulting in a diagnostic yield of 100%. The yield of mutation detection was 93% with three recessive and three sporadic cases not yielding a second mutation. Among these mutations, 13 in CLCN1 and 3 in SCN4A were novel. In conclusion, the current results show that in tandem analysis of CLCN1 and SCN4A affords high-level mutation ascertainment in families with NDM.
Similar content being viewed by others
Introduction
Non-dystrophic myotonias (NDMs) are a group of skeletal muscle disorders that have myotonia as their common feature, in reference to a delayed muscle relaxation after voluntary or evoked muscle contraction. Myotonic discharges can be recorded by needle-electromyography (needle-EMG) from the skeletal muscles of these patients. Clinically, NDMs are classified as dominant and recessive myotonia congenita (dominant MC; Thomsen's disease (OMIM 160800) and recessive MC; Becker's disease (OMIM 255700)), paramyotonia congenita (PC (OMIM 168300)) and potassium-aggravated myotonias (PAMs (OMIM 608390)). As suggested by Rüdel et al,1 PAM diagnosed without a potassium-loading test is referred to as a sodium-channel myotonia (SCM).
In 1971, Bryant and Morales-Aguilera showed that the membrane resistance of myotonic goat muscle fibres was considerably elevated at rest, which was found to be due to a strongly diminished sarcolemmal chloride conductance.2 The voltage-gated chloride channel concerned was shown to be involved in dominant and recessive MC in humans.3, 4 Subsequent genetic studies demonstrated a linkage to the skeletal muscle chloride channel gene (CLCN1 (OMIM 118425)), mapped to chromosome 7q35.5, 6 About 80 different CLCN1 mutations have, so far, been associated with MC.7 Meanwhile, in a second group of NDMs, impaired inactivation of voltage-gated sodium channels was observed.8 Various researchers later independently linked PC and PAM to the skeletal muscle sodium channel gene (SCN4A (OMIM 603967)), genetically mapped to chromosome 17q23–25.9, 10, 11 To date, at least 30 different missense mutations have been identified in this gene.12
Although many mutations have been identified in CLCN1, 25–60% of the MC patients who were examined lacked any identifiable CLCN1 mutation.13, 14, 15 Comparable studies in patients with PC or PAM have so far not been performed. Although sequencing of the entire CLCN1 increased the yield of putative myotonia-associated mutants, this was never able to account for all patients.16 Limitations in mutation detection methods, genetic heterogeneity and additional modifying factors were proposed to explain the discrepancy.13, 14, 15, 16 Plassart-Schiess et al17 postulated the incomplete dominance of some mutations with variable penetrance and expressivity as another compounding factor. Additionally, patients with suspected autosomal dominant MC may show SCN4A mutations. The purpose of the present study was therefore to optimize the genetic characterization of patients with NDM in The Netherlands by in tandem analysis of CLCN1 and SCN4A, as necessary.
Patients and methods
Proband selection
The current investigation comprised a cross-sectional, nationwide study. In March 2005, consultant neurologists across The Netherlands as well as the Vereniging Spierziekten Nederland (VSN), our national Patient Association for Neuromuscular Diseases, were requested to report patients with a clinical diagnosis of NDM to our research group over a full year. All patients were subsequently contacted and those who responded positively were invited to the neurology outpatient clinic of the Radboud University Nijmegen Medical Centre for the proposed clinical assessment, needle-EMG and collection of blood samples for genetic analysis. Inclusion criteria were age over 18 years, a clinical diagnosis of NDM according to established clinical criteria (Table 1),18 and myotonic discharges upon needle-EMG examination.
Exclusion criteria included a clinical or genetic diagnosis of types 1 or 2 myotonic dystrophy, a clinical or genetic diagnosis of primary periodic paralysis and unwillingness or inability to reduce or stop drug therapy for myotonia for the duration of the study. This latter criterion was added to optimize the clinical and electrophysiological evaluations of the myotonia. The study was approved by the Medical Ethics Committee of the Radboud University Nijmegen Medical Centre and all patients gave their written informed consent prior to their participation.
Preliminaries and procedure
CLCN1 and SCN4A were sequenced in tandem following our specially designed study strategy. The decision to sequence first CLCN1 or SCN4A was based on established clinical criteria, which were independently verified for each patient by two authors (JT and GD). Disagreement was resolved by discussion. When previously described mutations were identified in the analysis of the first gene, we did not proceed with sequencing the second gene. In case of a suspected recessive myotonia congenita in which we detected one mutation, we did not proceed with sequencing SCN4A. In case of novel mutations, their status and inheritance patterns were determined by clinical evaluations of first-degree relatives and by direct sequence analyses of their DNA. In addition, all novel missense mutations were screened in a control panel consisting of the DNA from 50 healthy Dutch individuals (100 chromosomes). In case no mutations were found in the first gene or novel mutations were not confirmed in first-degree relatives, we subsequently sequenced the second gene.
Mutation analysis
For each patient two 10-ml blood samples were collected in EDTA tubes. Genomic DNA was isolated from peripheral blood by the method of Miller et al19 at the Leiden University Medical Centre and subsequently screened for mutations by direct sequence analysis of CLCN1 and/or SCN4A. PCR analysis, purification of the PCR products (Millipore Multiscreen HTS PCR plates), sequencing (Big Dye Terminator Cycle Sequencer kit from Perkin-Elmer) and the final analysis (ABI3730) were performed as described previously with some minor modifications, indicated between brackets.20 The primer sets used for amplification of CLCN1 were made according to Lorenz et al21 and primer sets designed for amplification of SCN4A can be found at www.lumc.nl/4080/DNA/SCN4A.html.
Results
Study cohort
The recruitment procedure yielded a total of 113 probands, 23 of whom did not respond to initial contacts while 10 eventually refused participation without specifying their reasons. Nine were unable to participate due to transportation problems and three were unable to visit the hospital because of serious co-morbidity. Another 14 probands were excluded based on the following exclusion criteria: primary periodic paralysis (n=7), unwillingness/inability to reduce and stop drug therapy for myotonia (n=5) and no clinical diagnosis of NDM with absence of myotonic discharges by needle-EMG (n=2). Accordingly, 54 probands with a clinically and electrophysiologically supported diagnosis of NDM took part in this study. We, moreover, clinically and genetically examined 18 (20%) affected and 74 (80%) unaffected first-degree relatives of those probands for whom novel mutations or indistinct inheritance patterns were established. Table 2 shows the basic demographics for all 54 probands, and their 92 first-degree relatives.
Preliminaries and procedure data
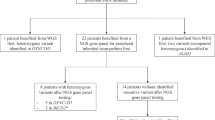
For an overview of the results, see the flowchart depicted in Figure 1. In short, CLCN1 was sequenced in 40 probands. For the remaining 14 probands, we first sequenced SCN4A. Mutations in CLCN1 were identified in 32 probands. Subsequently, the remaining eight showed a mutation in SCN4A. For all 14 probands in whom SCN4A was the first to be sequenced, a SCN4A mutation was found. DNA analysis thus identified 32 (59%) probands with CLCN1 mutations and 22 (41%) probands with SCN4A mutations, reflecting a 100% gene detection yield. However, the mutation detection yield, including homozygous recessives, was 93% (78 of 84) as in three recessive and three sporadic probands (worst case scenario) no second mutation was found.
Flowchart depicting the results of the in tandem analysis of CLCN1 and SCN4A. Flowchart presenting the numbers of probands (n, %) in whom the direct sequence analysis of CLCN1 or SCN4A was performed. The flowchart also presents the results. Note that based on established clinical criteria in 40 probands CLCN1 was sequenced first and that in 8 of these probands no mutation was identified. Also note that in 22 probands SCN4A was sequenced, 14 based on the mentioned clinical criteria and 8 based on the negative sequencing result of CLCN1. In total, CLCN1 mutations were identified in 32 families and SCN4A in 22.
Mutation analysis data
CLCN1 mutations
In 32 probands, 21 different CLCN1 mutations were identified, encompassing 9 missense, 5 splice site, 4 nonsense and 3 frameshift mutations (Table 3a). More than half (55%) of the mutations were detected in three exons (exons 3, 8 and 11). The remaining mutations were scattered across the entire chloride channel (Figure 2). Most of the 32 probands were compound heterozygote (n=17; Table 3b) and seven probands were homozygote, of whom four are known to result from consanguineous marriages. In eight probands, only one mutation was detected of whom two showed an autosomal dominant inheritance (families 37 and 51; Table 3b), three were sporadic (families 10, 16 and 38; Table 3b) and for the other three, autosomal recessive inheritance seemed plausible (families 9, 27 and 41; Table 3b). Overall, the F413C missense mutation was the most frequently observed mutation (n=8; see Table 3a).
The sites of the mutations in the skeletal muscle chloride (ClC-1) channel as identified in the present study. Novel mutations are underlined, circles represent dominant myotonia congenita (Thomsen) and squares recessive myotonia congenita (Becker). The membrane topology of the ClC-1 channel is adapted from Dutzler et al.45
In total, 13 of the 21 different mutations in CLCN1 were newly identified mutations (62%) and comprised two nonsense, four splice site, two frameshift and five missense mutations. The novel mutations 302−1G>A (n=4), S264fsX (n=4), M646I (n=3), 1167−10T>C (n=3), 302−2A>C (n=2) and A129T (n=2) were detected more than once in our families and none of the novel missense mutations were detected in the 100 control chromosomes.
Novel CLCN1 mutations
Y137X and C819X (recessive)
Two novel nonsense mutations were identified. The Y137X mutation was detected in a recessive pedigree (family 26). The C819X was identified as the only mutation in two affected sibs of family 41. The healthy mother appeared to be the carrier of this mutation.
K195fsX (recessive)
This novel frameshift mutation was identified in two patients of family 1 alongside the I479fsX mutation. A healthy sister, son and daughter appeared to be carriers of the K195fsX mutation.
S264fsX (recessive)
This mutation was detected in two patients of family 6 alongside the novel missense mutation G305E. The father was carrier of the S264fsX mutation and the mother and an unaffected brother were heterozygous for the G305E mutation. Furthermore, the S264fsX mutation was the only recessive mutation in the sporadic probands of families 10 and 16. Two unaffected children and three unaffected siblings of family 10 and the mother of proband 16 were carriers of this mutation. Lastly, S264fsX was also identified in family 43 alongside the novel splice-site mutation 1065−2A>C.
302−1G>A (recessive)
This novel splice-site mutation was identified in a compound heterozygous state in all three affected sibs of families 2, 4 and 5 and in the recessive pedigree of family 11.
302−2A>C (recessive)
This second novel splice-site mutation was observed in two families: homozygous in the proband of family 7, issue of a consanguineous marriage and compound heterozygous in a sporadic patient with a recessive mode of inheritance (family 54) alongside the newly identified missense mutation M646I.
1065−2A>C (recessive)
The third novel splice-site mutation was identified in the proband of family 43 alongside the novel frameshift mutation S264fsX. The mother and an unaffected sister were carriers of the novel splice-site mutation and the father and two siblings were carriers of the novel frameshift mutation.
1167−10C>T (recessive)
All affected sibs (n=5) of the recessive pedigrees 4 and 5 showed this novel splice-site mutation together with the novel 302−1G>A mutation. Unaffected first-degree relatives in both families appeared to be carriers of one of the novel mutations. This mutation was also identified in a compound heterozygous state in a single patient of family 14. First-degree unaffected relatives carried one of the two mutations.
A129T (dominant)
This missense mutation emerged in two families (37 and 38), ie one sporadic and one with a presumably dominant inheritance pattern. This could not be confirmed by DNA analysis because the affected father was deceased and the proband did not have children. SCN4A was also analysed retrospectively in both families and showed no mutations.
G305E (recessive)
The second novel missense mutation was identified in family 6 and occurred in all affected family members alongside a novel recessive frameshift mutation (S264fsX). The mutation was also identified in two unaffected siblings and the unaffected father.
E417G (recessive)
This mutation was detected in the proband (E417G/G285E) of family 40. The unaffected mother was heterozygous for the E417G mutation; the father was deceased.
K614M (recessive)
This fourth novel missense mutation was identified in family 42. The affected proband was compound heterozygous (K614M/180+3A>T). Both parents were deceased, while one brother was a carrier of the 180+3A>T and one sister was a carrier of the K614M mutation.
M646I (recessive)
The M646I mutation was identified in three different families (46, 48 and 54). It was identified alongside the recessive missense mutation G285E, the nonsense mutation R894X and the novel splice-site mutation 302−2A>C, respectively. In family 48, the mutation was also detected in two affected sibs. In the recessive pedigree of family 54, the described mutations occurred in a single patient. In family 46, the father was a carrier of G285E, and the mother was deceased.
Clinical features of the probands with CLCN1 mutations
All probands with CLCN1 mutations showed obvious clinical signs of myotonia and 97% showed the warm-up phenomenon. About 15% of the probands showed transient paresis and muscle wasting, 48% only showed transient paresis and 37% showed neither transient muscle weakness nor muscle wasting. The probands with novel mutations did not show new clinical features compared with those having already known mutations.
SCN4A mutations
In 22 probands, 11 different missense mutations were identified (Table 4). Three mutations were located in domain I, one in domain II and six in the domains III and IV of the voltage-gated sodium channel, Nav1.4 (Figure 3). In 59% of the probands, a mutation was identified in exon 22, of which the G1306V mutation was the most common (n=8).
The sites of the mutations in the skeletal muscle sodium (Nav1.4) channel as identified in the present study. Novel mutations are underlined, circles represent paramyotonia congenita and squares sodium channel myotonias. The membrane topology of the Nav1.4 channel is adapted from Jurkat-Rott et al.44
In total, 3 of the 11 different missense mutations were novel (27%), comprising two mutations in codon 250: L250V and L250P and one in codon 689: L689F. Each of the three novel mutations was detected once and none of the novel missense mutations were detected in the 100 control chromosomes. All probands with the novel mutations showed a phenotype mimicking Thomsen's disease. Therefore, DNA of these three probands was first sequenced for CLCN1 mutations, but no mutations were identified.
Novel SCN4A mutations
L250V (dominant)
This missense mutation in the proband of family 12 is most probably a de novo mutation. Presumably, both his parents (deceased) were unaffected while his daughter and granddaughter were both affected and showed the same L250V mutation.
L250P (dominant)
This mutation was identified in the proband of family 39. The affected father and brother in this family also showed this mutation. It was not detected in an unaffected brother and sister.
L689F (dominant)
This mutation was detected as a de novo mutation in family 36. Both parents of the proband were unaffected and did not carry the mutation. The mutation was identified in both the proband and her affected daughter.
Clinical features of the probands with SCN4A mutations
All probands with SCN4A mutations showed obvious clinical signs of myotonia, especially in the eyelid muscles. Furthermore, probands with SCM, including the three probands with novel mutations, showed the warm-up phenomenon. The phenomenon was detected in the eyelid muscles (80%) as well as in the hand flexor muscles (80%). In contrast, probands with paramyotonia congenita showed paramyotonia in the eyelid muscles (40%) as well as in the hand-flexor muscles (75%). Finally, almost all probands with paramyotonia congenita showed an increase of paramyotonia or a flaccid paresis after cooling. Probands with SCM did not react to cooling.
Discussion
By sequencing CLCN1 and SCN4A in tandem, we detected mutations in all our probands. The yield of mutation detection was 93%, with six cases (7%) not yielding a second mutation. This is a high percentage, especially compared to previous studies that identified CLCN1 mutations in 40 to 75% of their MC patients, whereas analysis of SCN4A was not included.13, 14, 15 Although the yield of our mutation detection was high, we failed to detect a second mutation in six probands. Possibly, deletions or other types of mutation deep in the intron or the promoter region of CLCN1 may underlie the disease in these cases.16
Our strategy yielded 13 novel mutations in CLCN1 and 3 in SCN4A. Although we did not perform in vitro studies, there are four lines of evidence affirming the suggestion that these mutations are pathogenic. First, 8 of the 13 novel mutations in CLCN1 were splice site, frameshift or nonsense mutations, which are predicted to eliminate channel function.27 Second, none of the novel missense mutations occurred in the 100 control chromosomes. Third, all original wild-type amino acids at the sites of the missense mutations were well conserved across chloride or sodium channels of different species and/or among human chloride or sodium channels, and fourth, all but one (A129) of the novel mutations were segregating with the disease.
Since the pathogenic status of missense mutations is less clear than the status of the other mutations, these mutations will individually be discussed by their location and conservation.
First, the A129T mutation is located in transmembrane segment B of the human ClC-1 channel. A129 is a highly conserved amino acid across ClC channels of different species and is well conserved among the human plasma membrane ClC isoforms ClC-1, -2, -Ka and -Kb. Furthermore, A129T is in the vicinity of the already established M128V and S132C mutations. Both mutations segregated with the Thomsen's phenotype.36, 37 M128V and S132C were both electrophysiologically characterized and showed a rightward shift in the current–voltage relationship, explaining their pathogenicity.36, 37, 38
Second, the G305E mutation is located in transmembrane segment 1 of ClC-1. G305 is a highly conserved amino acid across ClC channels of different species and is well conserved among the human ClC isoforms ClC-1, -2, -Ka and -Kb. G305E is in the vicinity of F307S, which was reported to drastically shift the voltage dependence of ClC-1 to positive potentials, preventing these channels from repolarizing muscle action potentials efficiently.24
Third, the E417G mutation is situated in the last codon of exon 11 and is therefore predicted to affect the splicing of this exon. However, further RNA studies are needed to explore this. Furthermore, E417, located in the linker between helix K and L of ClC-1, is highly conserved among ClC channels of different species.
Fourth, the K614M mutation, conserved across ClC channels of different species but not among human ClC isoforms, and the L646I mutation, conserved across ClC channels of different species and among the human isoforms ClC-1 and –2, are located in the β1 and β2–3 linker of the cystathionine β-synthase (CBS1) domain, respectively. Although the precise role of the CBS-domains is unknown, Estévez et al39 suggested that mutations in this domain will influence the voltage-gated dependence of gating through the common gate.
The L250P/V mutations in SCN4A are located in the membrane-spanning segment 5 of domain I of Nav1.4. Although in the vicinity of a benign polymorphism (S246L),40 L250 is highly conserved across Nav1.4 channels of different species and among the α-subunits of human sodium channels Nav1.1–Nav1.8. Furthermore, the mutations were retrospectively absent in 200 control chromosomes. Finally, both probands with these mutations showed a definite phenotype of NDM in the absence of other mutations in CLCN1 or SCN4A and both mutations were segregating with the disease.
The third novel SCN4A missense mutation (L689F), located in the linker between segments 4 and 5 of domain II in Nav1.4, is located at the same codon as the already established mutation L689I. This mutation was shown to cause in vitro effects of a hyperpolarizing shift in the voltage dependence of activation causing hyperkalemic periodic paralysis.41 The proband with the L689F mutation in our population showed a Thomsen-like phenotype without symptoms of weakness.
To exclude benign polymorphisms, we tested 100 control chromosomes in accordance with the current best practice guidelines. Furthermore, the Leiden University Medical Centre analysed approximately 500 patients with suspected NDM worldwide during the last 5 years and none of the detected variants were identified. Others mainly performed in vitro studies for the confirmation of novel mutations in one of these genes.36, 42, 43 In the future, such studies should also be done for the eight novel missense mutations detected in our study.
In this study, we identified 21 different CLCN1 and 11 different SCN4A mutations. Meyer-Kleine et al13 also found a high number of different mutations in their German-based cohort. In Scandinavian studies, three and eight different CLCN1 mutations were detected, respectively.16, 28 In one of these studies, the A513V, F413C and R894X mutations clearly predominated.16 Thus, our study yielded a broad spectrum of mutations underlying NDM in The Netherlands without clear evidence of a manifest founder effect. We hypothesize that these findings may be attributable to the high population density in The Netherlands, especially when compared to the low population densities in Sweden, Norway and Finland.
Our analyses revealed dominant MC to be scarce in The Netherlands, which is in sharp contrast with the initial clinical diagnoses. In 20% of the patients, the initial referral diagnosis was a dominant MC. However, only two of these patients were finally classified as dominant MC while the others proved to have SCM. All the probands with a SCM mutation showed a clinical picture of a generalized myotonia in combination with the warm-up phenomenon, mimicking Thomsen's disease.44
In conclusion, we have shown that in tandem analysis of CLCN1 and SCN4A affords a high level of mutation ascertainment in families with NDM. With this strategy, we were able to identify 13 novel CLCN1 and three novel SCN4A mutations. Moreover, it enabled us to confirm earlier suggestions that the prevalence of SCN4A mutations is higher than previously assumed.18 Based on the results presented here, we feel safe in suggesting that our approach shows great diagnostic potential and may offer optimal conditions for future genotype–phenotype studies.
References
Rüdel R, Ricker K, Lehmann-Horn F : Genotype–phenotype correlations in human skeletal muscle sodium channel diseases. Arch Neurol 1993; 50: 1241–1248.
Bryant SH, Morales-Aguilera A : Chloride conductance in normal and mytonic muscle fibres and the action of monocarboxylic aromatic acids. J Physiol 1971; 219: 367–383.
Rüdel R, Ricker K, Lehmann-Horn F : Transient weakness and altered membrane characteristic in recessive generalized myotonia (Becker). Muscle Nerve 1988; 11: 202–211.
Lipicky RJ, Bryant SH : A biophysical study of the human myotonias. In: Desmedt J (ed).: New Developments in Electromyography and Clinical Neurophysiology, Vol. 1 Basel: Karger, 1973, pp 451–563.
Koch MC, Steinmeyer K, Lorenz C et al: The skeletal muscle chloride channel in dominant and recessive human myotonia. Science 1992; 257: 797–800.
Abdalla JA, Casley WL, Cousin HK et al: Linkage of Thomsen disease to the T-cell-receptor beta (TCRB) locus on chromosome 7q35. Am J Hum Genet 1992; 51: 579–584.
Colding-Jørgensen E : Phenotypic variability in myotonia congenita. Muscle Nerve 2005; 32: 19–34.
Lehmann-Horn F, Rüdel R, Dengler R, Lorkovic H, Haass A, Ricker K : Membrane defects in paramyotonia congenita with and without myotonia in a warm environment. Muscle Nerve 1981; 4: 396–406.
Ebers GC, George Jr AL, Barchi RL et al: Paramyotonia congenita and hyperkalemic periodic paralysis are linked to the adult muscle sodium channel gene. Ann Neurol 1991; 30: 810–816.
Lerche H, Heine R, Pika U et al: Human sodium channel myotonia: slowed channel inactivation due to substitutions for a glycine within the III-IV linker. J Physiol 1993; 470: 13–22.
Ricker K, Moxley III RT, Heine R, Lehmann-Horn F : Myotonia fluctuans. A third type of muscle sodium channel disease. Arch Neurol 1994; 51: 1095–1102.
Cannon SC : Spectrum of sodium channel disturbances in the nondystrophic myotonias and periodic paralyses. Kidney Int 2000; 57: 772–779.
Meyer-Kleine C, Steinmeyer K, Ricker K, Jentsch TJ, Koch MC : Spectrum of mutations in the major human skeletal muscle chloride channel gene (CLCN1) leading to myotonia. Am J Hum Genet 1995; 57: 1325–1334.
Zhang J, George Jr AL, Griggs RC et al: Mutations in the human skeletal muscle chloride channel gene (CLCN1) associated with dominant and recessive myotonia congenita. Neurology 1996; 47: 993–998.
Sangiuolo F, Botta A, Mesoraca A et al: Identification of five new mutations and three novel polymorphisms in the muscle chloride channel gene (CLCN1) in 20 Italian patients with dominant and recessive myotonia congenita. Hum Mutat 1997; 11: 331.
Sun C, Tranebjaerg L, Torbergsen T, Holmgren G, Van Ghelue M : Spectrum of CLCN1 mutations in patients with myotonia congenita in Northern Scandinavia. Eur J Hum Genet 2001; 9: 903–909.
Plassart-Schiess E, Gervais A, Eymard B et al: Novel muscle chloride channel (CLCN1) mutations in myotonia congenita with various modes of inheritance including incomplete dominance and penetrance. Neurology 1998; 50: 1176–1179.
Lehmann-Horn F, Rüdel R : Non-dystrophic myotonias and periodic paralyses.In: Emery AEH (ed).: Diagnostic Criteria for Neuromuscular Disorders. London: Royal Society of Medicine Press, 1997, pp 31–36.
Miller SA, Dykes DD, Polesky HF : A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 1988; 16: 1215.
Ginjaar HB, van der Kooi AJ, Ceelie H et al: Sarcoglycanopathies in Dutch patients with autosomal recessive limb girdle muscular dystrophy. J Neurol 2000; 247: 524–529.
Lorenz C, Meyer-Kleine C, Steinmeyer K, Koch MC, Jentsch TJ : Genomic organization of the human muscle chloride channel CIC-1 and analysis of novel mutations leading to Becker-type myotonia. Hum Mol Genet 1994; 3: 941–946.
Sloan-Brown K, George Jr AL : Inheritance of three distinct muscle chloride channel gene (CLCN1) mutations in a single recessive myotonia congenita family. Neurology 1997; 48: 542–543.
George Jr AL, Sloan-Brown K, Fenichel GM, Mitchell GA, Spiegel R, Pascuzzi RM : Nonsense and missense mutations of the muscle chloride channel gene in patients with myotonia congenita. Hum Mol Genet 1994; 3: 2071–2072.
Kubisch C, Schmidt-Rose T, Fontaine B, Bretag AH, Jentsch TJ : ClC-1 chloride channel mutations in myotonia congenita: variable penetrance of mutations shifting the voltage dependence. Hum Mol Genet 1998; 7: 1753–1760.
Meyer-Kleine C, Ricker K, Otto M, Koch MC : A recurrent 14 bp deletion in the CLCN1 gene associated with generalized myotonia (Becker). Hum Mol Genet 1994; 3: 1015–1016.
Steinmeyer K, Lorenz C, Pusch M, Koch MC, Jentsch TJ : Multimeric structure of ClC-1 chloride channel revealed by mutations in dominant myotonia congenita (Thomsen). EMBO J 1994; 13: 737–743.
Pusch M : Myotonia caused by mutations in the muscle chloride channel gene CLCN1. Hum Mutat 2002; 19: 423–434.
Papponen H, Toppinen T, Baumann P et al: Founder mutations and the high prevalence of myotonia congenita in Northern Finland. Neurology 1999; 53: 297–302.
Rosenfeld J, Sloan-Brown K, George Jr AL : A novel muscle sodium channel mutation causes painful congenital myotonia. Ann Neurol 1997; 42: 811–814.
Sugiura Y, Aoki T, Sugiyama Y, Hida C, Ogata M, Yamamoto T : Temperature-sensitive sodium channelopathy with heat-induced myotonia and cold-induced paralysis. Neurology 2000; 54: 2179–2181.
Ptácek LJ, Tawil R, Griggs RC et al: Sodium channel mutations in acetazolamide-responsive myotonia congenita, paramyotonia congenita and hyperkalemic periodic paralysis. Neurology 1994; 44: 1500–1503.
McClatchey AI, Van den Bergh P, Pericak-Vance MA et al: Temperature-sensitive mutations in the III-IV cytoplasmic loop region of the skeletal muscle sodium channel gene in paramyotonia congenita. Cell 1992; 68: 769–774.
Yamada T, Ochi H, Hara H, Yoshimura T, Kobayashi T : A skeletal muscle sodium channel mutation in a Japanese family with paramyotonia congenita. J Neurol Sci 1995; 133: 192–193.
Ptácek LJ, George Jr AL, Barchi RL et al: Mutations in an S4 segment of the adult skeletal muscle sodium channel cause paramyotonia congenita. Neuron 1992; 8: 891–897.
Bulman DE : Phenotype variation and newcomers in ion channel disorders. Hum Mol Genet 1997; 6: 1679–1685.
Wu FF, Ryan A, Devaney J et al: Novel CLCN1 mutations with unique clinical and electrophysiological consequences. Brain 2002; 125: 2392–2407.
Grunnet M, Jespersen T, Colding-Jørgensen E et al: Characterization of two new dominant ClC-1 channel mutations associated with myotonia. Muscle Nerve 2003; 28: 722–732.
Colding-Jørgensen E, Dunø M, Schwartz M, Vissing J : Decrement of compound muscle action potential is related to mutation type in myotonia congenita. Muscle Nerve 2003; 27: 449–455.
Estévez R, Pusch M, Ferrer-Costa C, Orozco M, Jentsch TJ : Functional and structural conservations of CBS domains from CLC chloride channels. J Physiol 2004; 557: 363–378.
Tsujino A, Maertens C, Ohno K et al: Myasthenic syndrome caused by mutation of the SCN4A sodium channel. Proc Natl Acad Sci USA 2003; 100: 7377–7382.
Bendahhou S, Cummins TR, Kula RW, Fu YH, Ptácek LJ : Impairment of slow inactivation as a common mechanism for periodic paralysis in DIIS4-S5. Neurology 2002; 58: 1266–1272.
Simpson BJ, Height TA, Rychkov GY et al: Characterization of three myotonia-associated mutations of the CLCN1 chloride channel gene via heterologous expression. Hum Mutat 2004; 24: 185.
Bouhours M, Sternberg D, Davoine CS et al: Functional characterization and cold sensitivity of T1313A, a new mutation of the skeletal muscle sodium channel causing paramyotonia congenita in humans. J Physiol 2004; 554: 635–647.
Jurkat-Rott K, Lehmann-Horn F : Muscle channelopathies and critical points in functional and genetic studies. J Clin Invest 2005; 115: 2000–2009.
Dutzler R, Campbell EB, Cadene M, Chait BT, MacKinnon R : X-ray structure of a ClC chloride channel at 3.0 Å reveals the molecular basis of anion selectivity. Nature 2002; 415: 287–294.
Acknowledgements
We thank the VSN for their cooperation in this study, and Linda Vijfhuizen and Naima Lamzira for their expert technical assistance and G van Zonneveld for illustrating figures. This study was supported by Prinses Beatrix Fonds (MAR04-0118), The Netherlands.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Trip, J., Drost, G., Verbove, D. et al. In tandem analysis of CLCN1 and SCN4A greatly enhances mutation detection in families with non-dystrophic myotonia. Eur J Hum Genet 16, 921–929 (2008). https://doi.org/10.1038/ejhg.2008.39
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2008.39
Keywords
This article is cited by
-
Community data-driven approach to identify pathogenic founder variants for pan-ethnic carrier screening panels
Human Genomics (2023)
-
Non-dystrophic myotonias: clinical and mutation spectrum of 70 German patients
Journal of Neurology (2021)
-
Association of Three Different Mutations in the CLCN1 Gene Modulating the Phenotype in a Consanguineous Family with Myotonia Congenita
Journal of Molecular Neuroscience (2021)
-
Up-regulation of voltage-gated sodium channels by peptides mimicking S4-S5 linkers reveals a variation of the ligand-receptor mechanism
Scientific Reports (2020)
-
Clinical exome sequencing in neuromuscular diseases: an experience from Turkey
Neurological Sciences (2020)