Abstract
The triple A syndrome is caused by autosomal recessively inherited mutations in the AAAS gene and is characterized by achalasia, alacrima and adrenal insufficiency as well as progressive neurological impairment. We report on a 14-year-old girl with slowly progressive axonal motor neuropathy with conspicuous muscle wasting of hypothenars and calves as well as alacrima. The mutation analysis of the AAAS gene revealed a compound heterozygous mutation: a c.251G>A mutation in exon 2 that had been reported previously, and a novel c.1288C>T mutation in exon 14. At the transcriptional level, the c.251G>A transition results in an aberrant splicing and decay of this RNA strand so that the particular clinical picture results from the novel c.1288C>T, (p.Leu430Phe, L430F) mutation in a hemizygous form. With transfection experiments, we demonstrate that GFP–ALADINL430F correctly localizes to nuclear pore complexes. Therefore, we conclude that this point mutation impairs ALADIN function at the nuclear pore.
Similar content being viewed by others
Introduction
Triple A syndrome (MIM #231550) is a rare autosomal recessive inherited disease characterized by its three main symptoms alacrima, achalasia and adrenal insufficiency as well as neurological abnormalities.1 Triple A syndrome is caused by mutations in the AAAS gene on chromosome 12q13.2, 3 AAAS is ubiquitously expressed with an enhanced expression in the pituitary gland, the gastrointestinal tract and the adrenal gland, tissues that are mostly affected in triple A syndrome.
The protein encoded by the AAAS gene has been designated ALADIN (alacrima achalasia adrenal insufficiency neurologic disorder). With its four WD-repeat domains, it belongs to the family of the WD-repeat proteins. In a proteomic approach, Cronshaw et al.4 identified ALADIN as a new member of the nucleoporin family at the nuclear pore complex (NPC). Nucleoporins play crucial roles in transport processes between the nucleus and cytoplasm. Furthermore, they are involved in the regulation of cell cycle and gene expression.5 Although the precise function of ALADIN has not yet been elucidated, it was shown that the correct localization of ALADIN at the nuclear pore plays an important role for its function.6, 7 All naturally occurring pathogenic mutations investigated so far resulted in a mislocalization of the mutant ALADIN protein in the cytoplasm or in an even distribution between the nucleus and cytoplasm.
Nearly all patients with the three main features of triple A syndrome and proven AAAS mutations show additional neurological abnormalities comprising autonomic disturbances, optic atrophy and involvement of other cranial nerves, upper and lower motor neuron signs and mental retardation.8 These symptoms may appear successively over a period of many years. As patients show a variety of concomitant symptoms, the disease is regarded as a multisystemic disorder.
Here, we report the first AAAS mutation, which does not affect the correct targeting of ALADIN to the NPC suggesting a functional impairment of the mutant protein rather than an effect of false localization. In our 14-year-old patient, this mutation is associated with a slowly progressive axonal motor neuropathy with uncommon distribution of muscle wasting as the main presenting clinical feature for over a decade.
Patient and methods
Case report
This 14-year-old girl is the first child of healthy nonconsanguineous parents. The family history is unremarkable, one older brother of the patient is healthy. After an uneventful pregnancy, the patient was born at term without complications. Birth weight, length and head circumference were in the normal range, and her somatic growth was normal later on. She showed mild muscular hypotonia, she was able to sit without support at 9 months and walked freely at 18 months. Starting at about 3 years of age, a slowly progressive gait disturbance with abnormal posture of lower legs and feet as well as mild atrophy of calves were noted. Nerve conduction studies and somatosensory-evoked potentials were normal at that time.
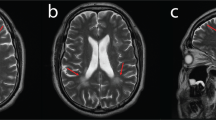
During the following years, a continuous slow deterioration with predominantly distal weakness and muscle wasting occurred. Muscular atrophy affected predominantly the hypothenars and the calves. At 9 years of age, she showed marked pes cavus and was able to walk on her heels, but not on her toes. Muscle stretch reflexes were normal in arms and legs. At this time, motor nerve conduction studies revealed marked reduction of compound muscle action potential amplitudes, whereas conduction velocities were normal or mildly reduced (Table 1). Sensory nerve conduction studies were normal. Electromyography (EMG) of the gastrocnemius muscle was considered myopathic. Muscle ultrasound showed unspecifically increased echogenicity consistent with both, myopathic and neurogenic changes in quadriceps, and predominantly myopathic alterations in gastrocnemic muscles. The magnetic resonance imaging (MRI) of the calves performed at 10 years of age demonstrated the predominant affection of gastrocnemic muscles (Figure 1). Histological and immunohistochemical investigations of a biopsy of the quadriceps muscle revealed variability in fiber size, striking fiber type I predominance and rare atrophic fibers without grouping (Figure 2e). These alterations were considered to indicate an unspecific but likely myopathic process without clear neurogenic features.
Axial MRI of the lower legs of (a) a healthy control and (b) the patient at 10 years of age without contrast agent. In a healthy control (a), the tibia is labeled with open white arrows. Calve muscles are depicted with normal volume and normal homogeneous signal intensity. In patient (b), marked atrophy of the calves with inhomogeneous signal intensity of muscle, fatty and connective tissue is visible (full white arrows).
At 15 years, muscle atrophy predominantly of (a) calves and (b) hypothenars (c) lingua plicata and hypertrophic papillae of the tongue as well as (d) plantar hyperkeratosis are apparent. (e) Biopsy of the quadriceps muscle at the age of 10 years, ATPase at pH 4.3 and × 20 magnification, shows variability in fiber size, striking fiber type I predominance and rare atrophic fibers without grouping (black arrow).
Physical examination at 14 years revealed distally pronounced muscular weakness and atrophy in the lower more than in the upper extremities with marked atrophy of the calves (Figure 2a) and hypothenars (Figure 2b). Muscle stretch reflexes in the arms and quadriceps reflex were exaggerated, ankle reflexes were absent. Plantar response was flexor. No sensory disturbance was found. The walking distance was at most 1000 m. Hand function was impaired. Lingua plicata and hypertrophic papillae of the tongue (Figure 2c) as well as hyperkeratosis of soles (Figure 2d) and knees were apparent. Nerve conduction studies confirmed an axonal motor neuropathy.
Additionally, a disturbance of tear production with the onset during her first months of life had been noted. Ophthalmological examination resulted in the diagnosis of sicca syndrome, and artificial tears were prescribed. Serologic investigations provided no evidence for Sjögren syndrome. Renewed discussion with the mother revealed that the production of tears was not reduced, but in fact has always been completely absent. This resulted in a reappraisal of this sign as being alacrima.
After AAAS mutation analysis had confirmed triple A syndrome, we specifically asked for any symptoms of achalasia and the girl reported mild swallowing difficulties. Esophageal manometry and barium esophagogram now revealed incipient achalasia. On the last follow-up, at age 17 years, an ACTH stimulation test still provided no evidence for adrenal insufficiency. Serum levels of cortisol were 169 nmol/l (normal range: 138–690 nmol/l) unstimulated and 648 nmol/l (normal range: >552 nmol/l) 60 min after i.v. administration of 250 μg ACTH. Basal ACTH serum level was 1.75 pmol/l (normal range: <11).
Molecular genetic and functional analysis
Blood sample from patient was collected after obtaining written informed consent for DNA analysis. DNA preparation was performed according to the standard protocols using the QIAamp DNA Mini Kit (QIAGEN Inc., Valencia, CA). Coding sequences of the 16 exons of AAAS, including exon–intron boundaries, were amplified from genomic DNA. Primer sequences and PCR conditions used for amplification are available upon request. Total RNA was prepared from patient and control EBV-transformed lymphocytes and fibroblasts using RNeasy Mini Kit (QIAGEN Inc.). After reverse transcription of mRNA with SuperScript™ III Reverse Transcriptase (Invitrogen, Karlsruhe, Germany), the sequences of exons 1–3 and exons 13–16 were amplified using the primers P1+P2 and P3+P4, respectively (Table 2).
The relative amount of AAAS mRNA in fibroblast cells was quantified using TaqMan® – PCR with the ABI PRISM® 7300 Sequence Detection System (Applied Biosystems, Foster City, CA, USA). Probes were labeled with the reporter dye 6-carboxyfluorescein (6′-FAM) at the 5′ end and with the quencher dye 6-carboxytetramethylrodamine (TAMRA) at the 3′ end. Primers and probes were designed using Primer Express® 2.0 software (Applied Biosystems). The PCR product spanned the borders of AAAS exon 7 and 8 junctions, so that a potential genomic DNA contamination was unable to amplify. The following primers were used: AAAS forward primer 5′-CCCTACCTCCTTGTCTACCCG-3′, AAAS reverse primer 5′- CAGGTGTATGCCCAGGGTG-3′ and AAAS probe 5′-(FAM)- TCTTCTGGCTGTGCCCAAGTGCTGTC-(TAMRA)-3′. Gene transcription was normalized in relation to the transcription of the housekeeping gene glyceraldehyde-3-phosphate-dehydrogenase (GAPDH) with following primers: GAPDH forward primer 5′-GAAGGTGAAGGTCGGAGTC-3′, GAPDH reverse primer 5′-GAAGATGGTGATGGGATTTC-3′ and GAPDH probe 5′-(FAM)-CAAGCTTCCCGTTCTCAGCC-(TAMRA)-3′. Quantitative PCR reaction was performed in triplicates of three different probes. For each PCR, 20 μl of a Mastermix containing 2 μl of cDNA, 15 mM Tris-HCl pH 8.0, 50 mM KCl, 1.5 mM MgCl2 (AAAS)/2 mM MgCl2 (GAPDH), 0.8 mM dNTPs, 0.5 μ M ROX Reference Dye (Invitrogen, Karlsruhe) 0.9 μmol of each primer, 0.2 μmol probe and 1 U of AmpliTaq Gold DNA polymerase (Applied Biosystems) were used. Thermal cycling conditions were as follows: 10 min at 95°C followed by 40 repeats of 15 s at 95°C and 1 min at 60°C.
To detect the expression of erroneous spliced RNA, two different approaches were used. For the detection of a potential integration of intron 2 into the splice product, a nested PCR was carried out using primers from exon 1 and intron 2 (P1, P5, P6 and P7; Table 2 and Figure 4b). After the first PCR with primers P1 and P5 (502 bp product), a nested PCR (224 bp product) with primers P6 and P7 was performed. For the detection of a potential splice product with exon 2 deletion, we designed a reverse primer composed of the last 4 bp of exon 1 and 14 bp of exon 3 (P8, Table 2) and used it in combination with the forward primer from exon 1 (P1). One microliter of cDNA were amplified in 20 μl of a Mastermix containing 15 mM Tris-HCl pH 8.0, 50 mM KCl, 1.5 mM MgCl2, 0.2 mM dNTPs, 0.3 μmol of forward and reverse primer and 1 U of AmpliTaq Gold DNA polymerase (Applied Biosystems). PCR conditions were as follows: 95°C denaturation for 30 s, 64°C annealing for 20 s and 72°C extension for 30 s with 40 cycles, and another 72°C extension for 5 min.
RNA analysis from patient and control cells. (a) Agarose gel electrophoresis of cDNA-derived PCR products using the primers amplifying exons 1–3 (306 bp, lanes 1–3) and exons 1 to the aberrant junction of exons 1 and 3 (141 bp, lanes 4–6). PCR-amplified cDNA fragments were analyzed by electrophoresis in a 2% agarose gel. Lanes 1 and 4 show PCR products of WT cDNA, lanes 2 and 5 show the amplicons of patients cDNA and lanes 3 and 6 show the PCR products of water controls, lane 7 shows a 100 bp ladder. (b) Schematic illustration of the normal and aberrant mRNA splicing variants of our patient including primer positions for PCR analysis. (c) AAAS mRNA expression analysis performed by TaqMan PCR.
PCR products were purified using QIAquick columns (QIAGEN Inc.) and sequenced on an ABI 3100 genetic analyzer using BigDye Terminator Cycle Sequencing Kit 1.1 (Applied Biosystems).
Transfection and immunostaining were performed as described previously.7
Results
Sequencing of the genomic DNA of the patient revealed a compound heterozygous mutation. We found a G>A transition at nucleotide position 251(c.251G>A), the last base of exon 2, and a novel C>T transition at nucleotide position 1288 in exon 14 (c.1288C>T) that results in an amino-acid change from leucin 430 to phenylalanine (p.Leu430Phe; Figure 3a). The family analysis showed that both mutations are located on different alleles in that the c.251G>A mutation occurred at the paternal allele and the c.1288C>T mutation on the maternal allele (Figure 3b).
Molecular genetic analysis: (a) sequence chromatograms of DNA analysis of the patient and a healthy control individual. The arrows indicate the nucleotide altered by the mutation. (b) Schematic illustration of the maternal and paternal allele of the AAAS gene and the consequences of these mutations on RNA level. Mutated exons are highlighted in gray. (c) Sequence chromatograms of mRNA analysis amplifying exons 1–3, exon 1 to intron 2 and exons 13–16 of the patient and a healthy control individual. The arrows indicate the nucleotide altered by the mutation.
RT-PCR analysis on RNA from patient and control cells showed that the mutation c.251G>A in exon 2 leads to a splicing defect with subsequent nonsense-mediated mRNA decay (Figure 4). The mutation c.251G>A leads either to the splicing of exon 1 directly into exon 3 with skipping of the entire exon 2 or to the lack of intron 2 splicing (Figures 3c and 4b). Using a primer pair from exons 1 and 3 on cDNA of the patient, only the WT allele could be amplified (Figure 3c, column 1 and Figure 4a, lane 2). The PCR product of 5420 bp including intron 2 was not amplified under these conditions and the 178 bp splicing product of exon 1 directly into exon 3 was hardly ever visible. However, the latter product could be amplified by a forward primer from exon 1 and a reverse primer, which spans the exons 1/3 boundary and could only bind to this aberrant product (Figure 4a, lane 5). By the use of a forward primer from exon 1 and a reverse primer from intron 2 in a nested PCR, only the mutant allele of the patient was amplified and therefore a hemizygous G>A transition was observed by sequencing the RT-PCR product of the patient (Figure 3c, column 2). The RT-PCR product of exon 1 to intron 2 could only be amplified by a nested PCR indicating that the aberrant splicing results in a nonsense-mediated mRNA decay of this paternally derived aberrant mRNA strand. Any remaining RNA will result in a truncated protein. The lack of intron 2 splicing results in a premature stop codon at amino-acid position 84 (TGG>TAG → Trp84X) and skipping of exon 2 results in a frameshift at amino-acid position 42 with a premature stop codon at amino-acid position 43 (Trp42AlafsX1) (Figure 4b). Results from AAAS–mRNA expression assay by TaqMan®-PCR disclosed that patient cells carry approximately half of the AAAS–mRNA compared with wild-type cells (Figure 4c). For this reason, in vivo ALADINL430F is most probably the only transcript deriving from this compound heterozygous mutation (Figure 3b and c, column 3).
Transfection experiments using a GFP–ALADIN construct in HeLa cells revealed that GFP–ALADINL430F like GFP–ALADINWT localizes to the NPCs (Figure 5). Hence, in contrast to other reported mutants,6, 7 the point mutation p.Leu430Phe does not impair the targeting of ALADIN to the NPCs.
HeLa cells were transfected with wild-type EGFP–ALADINWT or mutant EGFP–ALADINL430F. (a) Localization of GFP-tagged ALADIN constructs (green). (b) Localization of the nuclear pore complexes indicated by the anti-NUP62 antibody (red). (c) Colocalization of the green (EGFP–ALADIN) and the red (anti-NUP62) fluorescence is depicted by a yellow overlay of both fluorescences.
Discussion
The phenotype of triple A syndrome is highly variable. Molecular testing of the AAAS gene in our patient confirmed the diagnosis of triple A syndrome caused by two heterozygous point mutations. Whereas the first mutation c.251G>A was described previously, the second mutation c.1288C>T (p.Leu430Phe) is a novel mutation.3, 9 The first mutation results in an aberrant splicing with subsequent nonsense-mediated mRNA decay. This pathway of mRNA surveillance degrades transcripts containing premature termination codons.10 Therefore, the phenotype of the patient is probably dependent on the dysfunction of the ALADINL430F only. Intriguingly, functional testing of the novel p.Leu430Phe mutation revealed a correct localization of GFP–ALADINL430F at the NPC. Up to now, all investigated naturally occurring pathogenic AAAS mutations result in mislocalization of ALADIN away from the NPC to a predominantly cytoplasmic distribution.6, 7 Although the mutants ALADINQ15K and ALADINL25P were previously described as apparently remaining at the nuclear pore,7 ALADINL25P turned out to be a polymorphism and ALADINQ15K was discovered to be a missense mutation only in vitro, but in vivo results in aberrant splicing causing a frameshift followed by an early stop codon (p.G14fsX58).7 We conclude that the leucine 430 mutation does not interfere with the correct targeting of ALADIN to the NPC but may rather be critical for the interaction of ALADIN with other protein(s) of the nuclear pore or shuttling proteins and that an amino-acid substitution at this residue results in ALADIN dysfunction. This functional consequence of ALADINL430F could conceivably be different from other reported mutations, which result in ALADIN mislocalization thereby causing a particular course of the disease perhaps without adrenal insufficiency.
Before the discovery of AAAS mutations as the molecular cause for the triple A syndrome, it had already been pointed out that the significant neurological problems are common in this multisystem disorder.11 In a series of 20 families with clinical features of triple A syndrome, a wide spectrum of associated neurological abnormalities in all patients with triple A phenotype with and without proven AAAS mutations has been reported.8 The progressive neurological syndrome observed in eight patients from six families comprised pupillary and other cranial nerve abnormalities, optic atrophy, autonomic neuropathy as well as upper and lower motor neuron signs. Lower limb amyotrophy was noted, but the pattern of muscle involvement was not mentioned. Peripheral nerve involvement was characterized by a predominant motor axonal neuropathy with severe selective ulnar nerve affection.8
In our patient, a neuromuscular disorder with slowly progressive muscle wasting, predominantly of the hypothenars and the calves, was the main presenting clinical feature for over a decade with imperceptible onset during the first years of life. Nerve conduction studies clearly indicated a motor axonal neuropathy, but EMG, muscle ultrasound, muscle MRI and histological analysis of a muscle biopsy raise the possibility of additional myopathic features in this patient. To our knowledge, this has not been observed previously in triple A syndrome. However, the myopathic features seen in the biopsy were quite nonspecific. As the biopsy was from the relatively unaffected quadriceps muscle and not from the clinically and radiologically most affected gastrocnemius, a final determination whether there could be a myopathic component in addition to the definite axonal neuropathy has to remain open but should be considered in future patients.
The distribution of muscle atrophy with a predominance of calve atrophy is uncommon in hereditary neuropathies. Charcot Marie Tooth neuropathies are characterized by early paresis of foot dorsiflexion leading to foot drop, which was well preserved in our patient, whereas she was unable to walk on her toes early in the course.
A disturbance of tear production had been recognized by the mother already in the first months of life. A reappraisal of this symptom as being representative of alacrima and not sicca syndrome provided the clinical clue that prompted us to perform the mutation analysis of the AAAS gene. It is generally recognized that alacrima is the earliest and most consistent clinical feature of triple A syndrome.8, 11, 12 Only after genetic confirmation of the diagnosis, when we specifically asked for additional typical symptoms, our patient reported mild swallowing difficulties, and an incipient achalasia was detected. Whether the p.Leu430Phe mutation is associated also with adrenal insufficiency can not be fully predicted at this point. In a recent investigation of the patient at 17 years of age, adrenal function has been normal, but manifestation of adrenal insufficiency later in life cannot be excluded and a close follow-up is required.
Our observation demonstrates that oligosymptomatic variants of triple A syndrome may occur with motor axonal neuropathy, distal weakness and alacrima as sole symptoms. The statement that it is important to inquire about tear production and swallowing difficulties in obscure neurological syndromes associated with axonal neuropathy should be reinforced on the basis of our case.13 Thus, mutation analysis of the AAAS gene should be considered in patients with progressive neurological abnormalities who show even only one or two of the classical clinical features of triple A syndrome, especially when a motor neuropathy with predominant wasting of hypothenars and calves is present.
References
Allgrove J, Clayden GS, Grant DB, Macaulay JC : Familial glucocorticoid deficiency with achalasia of the cardia and deficient tear production. Lancet 1978; 8077: 1284–1286.
Tullio-Pelet A, Salomon R, Hadj-Rabia S et al: Mutant WD-repeat protein in triple-A syndrome. Nat Genet 2000; 26: 332–335.
Handschug K, Sperling S, Yoon SJ, Hennig S, Clark AJ, Huebner A : Triple A syndrome is caused by mutations in AAAS, a new WD-repeat protein gene. Hum Mol Genet 2001; 10: 283–290.
Cronshaw JM, Krutchinsky AN, Zhang W, Chait BT, Matunis MJ : Proteomic analysis of the mammalian nuclear pore complex. J Cell Biol 2002; 158: 915–927.
Weis K : Regulating access to the genome: nucleocytoplasmic transport throughout the cell cycle. Cell 2003; 112: 441–451.
Cronshaw JM, Matunis MJ : The nuclear pore complex protein ALADIN is mislocalized in triple A syndrome. Proc Natl Acad Sci USA 2003; 100: 5823–5827.
Krumbholz M, Koehler K, Huebner A : Cellular localization of 17 natural mutant variants of ALADIN protein in triple A syndrome – shedding light on an unexpected splice mutation. Biochem Cell Biol 2006; 84: 243–249.
Houlden H, Smith S, De Carvalho M et al: Clinical and genetic characterization of families with triple A (Allgrove) syndrome. Brain 2002; 125: 2681–2690.
Huebner A, Kaindl AM, Knobeloch KP, Petzold H, Mann P, Koehler K : The triple A syndrome is due to mutations in ALADIN, a novel member of the nuclear pore complex. Endocr Res 2004; 30: 891–899.
Khajavi M, Inoue K, Lupski JR : Nonsense-mediated mRNA decay modulates clinical outcome of genetic disease. Eur J Hum Genet 2006; 14: 1074–1081.
Grant DB, Barnes ND, Dumic M et al: Neurological and adrenal dysfunction in the adrenal insufficiency/alacrima/achalasia (3A) syndrome. Arch Dis Child 1993; 68: 779–782.
Moore PS, Couch RM, Perry YS, Shuckett EP, Winter JS : Allgrove syndrome: an autosomal recessive syndrome of ACTH insensitivity, achalasia and alacrima. Clin Endocrinol (Oxf) 1991; 34: 107–114.
Ouvrier R, McLeod JG, Pollard JD (eds).: Peripheral Neuropathy in Childhood. Mac Keith Press: London, 1999, p 239.
Cai F, Zhang J : Study of nerve conduction and late responses in normal Chinese infants, children, and adults. J Child Neurol 1997; 12: 13–18.
Acknowledgements
We thank Dana Landgraf for the excellent technical assistance. This work was supported by a grant of the Deutsche Forschungsgemeinschaft (DFG, HU395/3-5) to AH.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Koehler, K., Brockmann, K., Krumbholz, M. et al. Axonal neuropathy with unusual pattern of amyotrophy and alacrima associated with a novel AAAS mutation p.Leu430Phe. Eur J Hum Genet 16, 1499–1506 (2008). https://doi.org/10.1038/ejhg.2008.132
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2008.132
Keywords
This article is cited by
-
Biallelic Variants in the Nuclear Pore Complex Protein NUP93 Are Associated with Non-progressive Congenital Ataxia
The Cerebellum (2019)
-
Triple A syndrome: two siblings with a novel mutation in the AAAS gene
Hormones (2019)
-
Long-term clinical follow-up and molecular genetic findings in eight patients with triple A syndrome
European Journal of Pediatrics (2012)
-
Neurological features in adult Triple-A (Allgrove) syndrome
Journal of Neurology (2012)
-
Two siblings with triple A syndrome and novel mutation presenting as hereditary polyneuropathy
European Journal of Pediatrics (2011)