Abstract
Previously we have localized to chromosome 3q21–q24, a predisposition locus for colorectal cancer (CRC), through a genome-wide linkage screen (GWLS) of 69 families without familial adenomatous polyposis or hereditary non-polyposis CRC. To further investigate Mendelian susceptibility to CRC, we extended our screen to include a further GWLS of an additional 34 CRC families. We also searched for a disease gene at 3q21–q24 by linkage disequilibrium mapping in 620 familial CRC cases and 960 controls by genotyping 1676 tagging SNPs and sequencing 30 candidate genes from the region. Linkage analysis was conducted using the Affymetrix 10K SNP array. Data from both GWLSs were pooled and multipoint linkage statistics computed. The maximum NPL score (3.01; P=0.0013) across all families was at 3q22, maximal evidence for linkage coming from families segregating rectal CRC. The same genomic position also yielded the highest multipoint heterogeneity LOD (HLOD) score under a dominant model (HLOD=2.79; P=0.00034), with an estimated 43% of families linked. In the case–control analysis, the strongest association was obtained at rs698675 (P=0.0029), but this was not significant after adjusting for multiple testing. Analysis of candidate gene mapping to the region of maximal linkage on 3q22 failed to identify a causal mutation. There was no evidence for linkage to the previously reported 9q CRC locus (NPL=0.95, P=0.23; HLODdominant=0.40, HLODrecessive=0.20). Our findings are consistent with the hypothesis that variation at 3q22 contributes to the risk of CRC, but this is unlikely to be mediated through a restricted set of alleles.
Similar content being viewed by others
Introduction
Family history is well established to be one of the strongest risk factors for the development of colorectal cancer (CRC).1 Although germline mutations in APC, the mismatch repair (MMR) genes, MUTYH/MYH, SMAD4, ALK3 and STK11/LKB1 are associated with Mendelian susceptibility to CRC, collectively, these do not account for the observed familial aggregation of the disease.2, 3 Direct evidence for uncharacterized high/moderate-penetrance CRC genes is provided by CRC families showing evidence against linkage to known loci and by kindreds that fulfil the clinical (Amsterdam) criteria for a diagnosis of hereditary non-polyposis colorectal cancer (HNPCC) but whose CRCs do not display MMR deficiency.4 Familial CRC risks in relatives of colorectal adenoma (CRA) cases parallel those seen in relatives of CRC cases.1 It is therefore likely that a significant proportion of inherited predisposition to CRC is mediated through susceptibility to CRAs. These observations have provided a strong rationale for searching of novel predisposition genes through genome-wide linkage screens (GWLSs) of hereditary non-FAP/HNPCC CRC families (hereditary non-syndromic colorectal cancer – HNSCRC).
Using a high-density SNP array, we have previously performed a GWLS of 69 HNSCRC families in which involvement of known predisposition genes had been excluded and a novel CRC susceptibility locus at 3q21–q24 was identified.5 Other workers have proposed additional loci for CRC susceptibility genes on the basis of linkage, most notably on 9q22.2–31.2.6, 7
To further examine the impact of unknown moderate/high-penetrance genes on CRC risk, we conducted a further GWLS on an additional 34 HNSCRC families using the same analytical platform employed in our first analysis. We then pooled data from scans. In an effort to predict the most likely location of the putative CRC gene on 3q21–q24, we undertook linkage disequilibrium (LD) mapping at 1676 tagging SNPs in 620 HNSCRC cases and 961 controls. In addition, we have sought to identify disease-causing variants by direct mutational analysis of candidate genes localizing to the region of maximal linkage on 3q22.
Materials and methods
Ascertainment and collection of families and cases
For clarity, we refer to our previously reported GWLS of 69 pedigrees as phase 18 and the current analysis of 34 pedigrees as phase 2. As before, familial CRC cases were ascertained through the COloRectal tumour Gene Identification (CORGI) Study Consortium.8 Briefly, pedigrees informative for linkage had at least three affected individuals (confirmed by pathology reports). Individuals were classed as affected if they had CRC at age ≤75 years or ‘significant’ adenomas (three or more synchronous or metachronous, and/or villous morphology, and/or severe dysplasia, and/or diameter >1 cm and/or presentation at ≤45 years of age). No patient had clinical features of Peutz-Jeghers syndrome (PJS), juvenile polyposis (JPS), hereditary mixed polyposis (HMPS) or inflammatory bowel disease. Germline MMR gene mutations were excluded by microsatellite instability (MSI) testing of BAT25 and BAT26 in two CRCs from each family; kindreds in which both cancers were unstable were excluded. Where one MSI+tumour was found or if the only available CRC was unstable, direct mutation screening of all coding regions of MSH2 and MLH1 was undertaken. In addition, all families with an attenuated familial adenomatous polyposis (FAP)-like phenotype (>5 adenomas) were tested for mutations in APC, and the entire data set was tested for the two common MYH variants (Y165C and G382D) using fluorescence-SSCP analysis.
The case–control study was based on 620 CRC cases (280 male, 340 female) with at least one affected first-degree relative ascertained through CORGI. As with the linkage families, none had clinical features of FAP, PJS, JPS, HMPS or inflammatory bowel disease. In addition, none had a family history compatible with a diagnosis of HNPCC. Samples collected from unaffected spouses/partners of cases served as a source of 965 controls (439 male, 526 female).
Genomic DNA was extracted from whole blood using the Chemagic Magnetic Separation Module 1™. All samples were obtained with informed consent and local ethical review board approval in accordance with the tenets of the Declaration of Helsinki.
Genome-wide linkage screen
The current analysis was based on 34 families segregating CRC and/or CRA in which involvement of known loci had been excluded. A GWLS of families was undertaken using the GeneChip® Mapping 10K Xba 142 Array (Affymetrix Inc., Santa Clara, CA). SNP genotypes were obtained by following the Affymetrix protocol. Briefly, for each sample, 250 ng of genomic DNA was digested with the restriction endonuclease XbaI for 2.5 h, mixed with Xba adapters and ligated using T4 DNA ligase for 2.5 h. Ligated DNA was added to four separate PCRs, cycled, pooled and purified to remove unincorporated ddNTPs. The purified PCR products were then fragmented and labelled with biotin-ddATP. Biotin-labelled DNA fragments were hybridized to the arrays for 18 h in an Affymetrix 640 hybridization oven. After hybridization, arrays were washed, stained and scanned using an Affymetrix Fluidics Station FS450 with images obtained using an Affymetrix GeneChip® 3000 scanner. Affymetrix GCOS software (v1.4) was used to obtain raw microarray feature intensities (RAS scores). RAS scores were processed using Affymetrix GTYPE (v4.0) software to derive SNP genotypes (Affymetrix Inc.).
Non-Mendelian error checking of genotypes and generation of Linkage format files from raw Affymetrix array (chp) files were performed using the program ProgenyLab (Progeny Inc., South Bend, IN). The map order and distances between SNP markers were based on the UCSC Human Genome browser. The program MERLIN (v0.10.1)9 was employed to search for additional unlikely genotypes consistent with potential genotyping errors. Data from this analysis were pooled with phase 1 data. The presence of LD between markers has the potential to inflate multipoint linkage statistics when founders are not genotyped. We therefore calculated the pair-wise r2 between consecutive pairs of SNP markers. A threshold of 0.16 for r2 has been advocated to define high-LD SNPs, the inclusion of which will distort linkage statistics.10 The influence of LD on linkage statistics was removed by considering each set of marker in LD (defined as sets in which each consecutive marker pair in the set had r2>0.16) and retaining the most centrally positioned SNP from each set.
Multipoint linkage analysis was undertaken by implementation of the program SNPLINK,11 which performs fully automated non-parametric (mode-of-inheritance free) and parametric analyses before and after LD removal by incorporation of the MERLIN (v0.10.1)9 and ALLEGRO (v1.1)12 programs, respectively. Parametric linkage in the presence of heterogeneity was assessed using heterogeneity LOD (HLOD) scores. HLOD scores and their accompanying estimates of the proportion of linked families (α) were calculated using the statistical software package ALLEGRO.
We derived LOD scores under both dominant and recessive models of inheritance with reduced penetrance and four liability classes dependent upon age at diagnosis (<50, 50–59, 60–69 and ≥70 years) based on models derived from segregation analysis.2 Initial analysis was based on CRC only. Disease allele frequencies were 0.017 under the dominant model and 0.183 under the recessive model. For the dominant model, penetrances were set at 0.044, 0.105, 0.213 and 0.420, with corresponding phenocopy rates of 0.0004, 0.002, 0.007 and 0.030. For the recessive model, penetrances were set at 0.054, 0.146, 0.331 and 0.638 with corresponding phenocopy rates of 0.00004, 0.0003, 0.0026 and 0.023.
A second analysis was undertaken in which individuals were classed as affected if they had either CRC or ‘significant’ CRA(s), as defined above. In the absence of robust data on the age-specific prevalence or incidence rates for CRA, CRAs were considered to be equivalent to CRC 15 years later, based on the date of the patient fulfilling the study criteria. This assumption of equivalence follows from data estimating the risk of malignant transformation,13 and we have used this in our previous analysis. In all analyses, unaffected individuals were considered uninformative (that is, of unknown phenotype).
Heterogeneity LOD scores follow a complex statistical distribution, which can be approximated by the maximum of two independently distributed variables. To obtain significance estimates for HLODs, these were first converted to a χ2, where χ2=2 loge 10 × HLOD and significance values (p1) were then derived, using the χ2 distribution with one degree of freedom. The nominal P-value for the HLOD score is then given by 0.5 × [1−(1−p1)(1−p1)].14
Multipoint non-parametric linkage analyses were performed using the SALL statistic generated by MERLIN. Results are reported in terms of an NPL statistic and its associated one-sided P-value. Under the null hypothesis of no linkage, the NPL statistic is distributed asymptotically as a standard normal random variable. For each analysis, we also calculated empirical genome-wide significance levels for the non-parametric NPL linkage statistics and LOD score (after markers in high LD were removed) using 10 000 simulations. At each of the iterations, we used ALLEGRO to simulate genotype data, using the original phenotypes, allele frequencies, marker spacing and missing data patterns. MERLIN9 was used to estimate information content (IC) for each chromosome provided by the marker set by use of the entropy information measure.
Familial risk of CRC attributable to linked regions
The familial risk of CRC attributable to linked regions in siblings, λs, was determined from allele-sharing probabilities between affected relative pairs.15 Bootstrapping was employed to derive 95% confidence intervals for λs.
Case–control analysis
Genotyping of samples was performed using customized Illumina Bead Arrays according to the manufacturer's protocols. DNA samples with GenCall scores <0.25 at any locus were considered ‘no calls’. A DNA sample was deemed to have failed if it generated genotypes at lesser than 95% of loci. A SNP was deemed to have failed if lesser than 95% of DNA samples generated a genotype at the locus. To ensure quality of genotyping, a series of duplicate samples were genotyped and cases and controls were genotyped in the same batches. Conversion of genotype data into formats suitable for processing was performed using in-house Perl scripts. All other statistical manipulations described were undertaken in S-Plus (version 7; Insightful, Com) or R (version 2.4.0).
Genotypic frequencies in control subjects for each SNP were tested for departure from Hardy–Weinberg equilibrium (HWE) using a χ2 test or Fisher's exact test in which an expected cell count was <5. The association between SNP genotype and risk of CRC was assessed by means of test statistics based on allele counts.
Copy number change analysis and mutational analysis
A search for copy number changes within 3q21–q24 was undertaken using data generated from Illumina Arrays. SNP GenCall scores were analysed using Illumina Bead Studio software. Visualization and analysis of genotype data from Illumina arrays for CNV was performed using BeadStudio software version. 3.0.27. Allelic signal intensity ratios and log R ratios were calculated and genome profiles generated using the Illumina Genome Viewer and Illumina Chromosome Browser 3.0.27.
Mutational analysis was conducted by bidirectionally sequencing amplified PCR fragments using BigDye Terminator chemistry on an ABI 3730 × l sequencer (Applied Biosystems, CA, USA). PCR primers were designed using Primer 3 software from genome contig sequences (NT_005612, NCBI Build 36.1). Primers were designed to facilitate investigation of all exons, intron–exon boundies and 5′- and 3′-UTR regions of genes (details available on request). Resultant traces were aligned and compared with the gene consensus sequences obtained from the human genome database using the software package Mutation Surveyor (Version 3.0; SoftGenetics, PA, USA).
Results
Description of families analysed for linkage
Within the families included in phase 1 (n=69) and phase 2 (n=34), there were 487 affected individuals, of whom 206 had a diagnosis of CRC (with or without adenoma) and 232 had ‘significant’ adenomas. The number of affected persons (CRC or significant CRA) per family ranged from 2 to 10, and the number of affected persons per family with DNA available ranged from 2 to 7. Ten of the 103 families contained affected persons in three generations, whereas 59 pedigrees contained affected family members in two generations. In the remaining pedigrees, affected family members were confined to a single generation (Table 1). A higher proportion of families in phase 1 contained affected individuals in multiple generations than in phase 2 (Table 1). The difference in composition of families between the two phases was not a consequence of pre-defined criteria for family collection, but simply the result of ascertainment potential through clinical genetic centres subsequent to phase 1.
The mean age at diagnosis of CRC in the families was 56 years, significantly less than the mean value of 70 years for age at diagnosis observed in the general white UK population. The mean age at diagnosis of CRA in the families was 50 years. The minimum age of diagnosis of CRC within each family ranged from 28 to 82 years (median 56 years).
Linkage analysis in the full sample set
A total of 121 Affymetrix 10K142.2 arrays were processed in phase 2. A number of parameters were employed throughout the study to determine data quality, and all genotypes were housed within the pedigree-storage program ProgenyLab. The average SNP call rate per array for phase 2 was 98.1% compared with 98.2% for phase 1. For DNA extracted from male subjects, it was possible to examine the 309 markers on the X chromosome for errors due to miscalls or PCR contamination. No SNPs were heterozygous in male samples. Two hundred and seventy-three markers were fixed or were without a single map location leaving 9646 usable SNPs (97.3%) of which 9337 mapped to autosomes. After removal of SNPs in LD, 7413/9646 (76.8%) markers remained. Less than 0.4% of the total SNP genotypes generated were considered unlikely by analysis with Progeny Lab and/or MERLIN. All such genotypes were removed from further analyses.
Multipoint non-parametric linkage analysis of all 103 families with and without the high-LD SNPs is shown in Figure 1a. The panels within Figure 1 confirm that inclusion of high-LD SNPs in the analysis can lead to inflated linkage statistics, but in most cases, the overall profile of the linkage statistics remained the same. Genome-wide mean IC scores were almost identical whether or not high LD were included, in phase 1, phase 2 and the combined data set (combined data set: 0.717 before and 0.712 after LD removal).
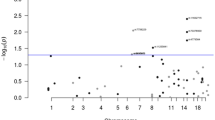
Table 2 details the highest NPL and HLOD scores after removal of SNPs in LD, for all autosomes in phases 1 and 2 separately, and in the combined data set. In the whole set of families, the strongest evidence of linkage (Figure 1) was for two chromosomal regions: 2p22, maximal under a recessive model (NPL=1.72; recessive HLOD=2.68); 11p15 (maximum NPL=2.25). The 12.3 Mb region supportive of linkage on 2p22 (recessive HLOD>1.0) was bounded by the SNPs rs954028 and rs1550429. There was no evidence for linkage to the previously reported locus between D9S971 and D9S272 on 9q22.33 (maximum NPL=0.4, P=0.34; HLODdominant=0.1; HLOD recessive=0.6).
Linkage analysis in CRC cases only
We then restricted our definition of affection status to CRC only, as, as we have argued above, separate loci might be associated with adenoma predisposition and progression to CRC. Restricting the affection status in this way rendered a subset of 50 families informative for linkage. The results for the whole genome are shown in Figure 2. The most robust evidence for linkage was attained at 3q22, under a dominant model (HLOD=2.79; maximum NPL=3.0). The dominant HLOD was maximized with 43% of families linked. The region of linkage (dominant HLOD>2.0) was bounded by SNPs rs1376949 and rs1427781, a distance of 7 Mb. Twenty-three of the 50 CRC families accounted for the majority of the linkage signal (NPL=3.03; HLOD=2.44). In the full data set, the 3q22 region produced an NPL score of 1.44 (Table 2). Intriguingly, of the 57 affected individuals in the nominally linked families from whom we had site of tumour data, 36 were affected with rectal disease (63%), a higher proportion than that expected on the basis of the relative frequency of colonic to rectal disease in the UK population (P<0.001).
Chromosomes 9p, 3p24, 13q31 and 17q24 displayed NPL scores suggestive of linkage on the basis of a 1% threshold, with NPL scores of 2.13, 3.01, 2.15 and 2.49, respectively (Table 3). In addition, there was weaker evidence of linkage to chromosome 2. Chromosomes 2p22 and 11p15 did not, however, attain the thresholds for suggestive linkage in the CRC-only data set. Figure 3 shows multipoint NPL and HLOD scores for the chromosomal region 3q22.
As with the analysis in the full sample set, there was no evidence in the CRC cases for linkage to the previously reported locus at 9q22.33 (maximum NPL=0.75, P=0.23; HLODdominant=0.40; HLOD recessive=0.20).
Contribution of the chromosome 3q21–q24 locus to the familial risk of CRC
The best estimate of the proportion of sibling pairs affected with CRC sharing no haplotype at chromosome 3q22 region was 0.12. This translates to a sibling relative risk attributable to the identified locus as 2.0 (95% confidence interval 1.3–3.7).
Association between 3q21–q24 tagging SNPs and colorectal cancer risk
Genotypes were obtained for 620 of 665 cases (93.2%) and 960 of 965 controls (99.5%). SNP call rates per sample were >99.6% in cases and controls. Of the 1676 SNPs, 96 were found to violate HWE in controls at the 5% significance level (expected number of failures, 84), thereby providing little evidence of population stratification or large-scale genotyping errors. After Bonferroni correction, four SNPs still violated HWE and were removed, leaving 1672 for further analysis. Each of the four SNPs removed had low genotyping reliability scores. There was a statistically significant association with CRC (P<0.05) for only nine of the 1230 SNPs. The most significant association was defined by rs698675 (P=0.0029), which maps to 141.8 Mb. None were, however, significantly associated with CRC risk after adjustment for multiple testing.
Analysis of copy number variation and mutational analysis of candidate genes
One affected individual with CRC from eight of the families that provided the best evidence of linkage to 3q22 was screened for CNV changes within the region. No CNVs within the extended region of linkage (10 Mb) on 3q21–24 were identified.
Using DNAs from 83 individuals belonging to the 69 phase 1 families displaying evidence of linkage to 3q22, we sequenced 30 candidate genes selected on the basis of known or predicted function, tissue-specific expression or position within the region of maximal linkage. Of these 30 genes, 23 mapped within the 4.4 Mb region of maximum linkage on 3q22 (Figures 3 and 4). Mutational analyses were also extended to include five genes positioned in the near proximity (MRPS22, NMNAT3, TOPBP1, RYK and AMOTL2) and to include MBD4 and GSK3b; two genes that represent very attractive candidates for CRC predisposition on the basis of biological interactions with known CRC susceptibility genes.
Schematic representation of the minimal region of linkage on chromosome 3q22.1–22.3. The genes mapping within the 4.4 Mb region are annotated with arrows positioned accordingly to their physical location in the genome. Mutational analyses by direct sequencing was performed for all genes within the region.
Information derived from the program SUSPECTS,16 which assesses the functional and sequence annotation similarity of the candidates with known disease predisposition genes, in combination with the expression profiles of each gene in normal tissue and CRC, was used to prioritize mutational analyses (Supplementary Table 1). The genes screened were MBD4, FNDC6, EPHB1, NCK1, PIK3CB, PPP2R3A, TOPBP1, STAG1, PCCB, CLDN18, FAIM, MRAS, FAM62C, CEP70, SOX14, A4GNT, TMEM22, ARMC8, FNDC6, MSL2L1, KY, DZIP1L, DBR1, MRPS22, NMNAT3, ANAPC3, RYK, AMOTL2, CEP63 and TXNDC6 (Supplementary Table 2). In addition to 101 documented polymorphisms, 95 novel changes were identified. These included 24 intronic changes, 26 synonymous changes and 45 non-synonymous changes (Supplementary Table 3). A single nonsense mutation was identified, in A4GNT, R266X, which leads to the truncation of the terminal 74 amino acids of the expressed protein. In all cases, pathogenicity could be excluded on the basis of type of sequence change, failure of the variant to segregate with disease haplotype or similarity in frequency between cases and healthy controls (Supplementary Table 3).
Discussion
The genetic basis of HNSCRC, or what has recently euphemistically been dubbed as ‘Syndrome X’, is presently ill-defined. On the basis of a GWLS of 74 sibling pairs affected with CRC or advanced adenomas from 53 CRC kindreds, a dominantly acting predisposition locus mapping to chromosome 9q22.2–q31 was reported by Wiesner et al6 to contribute to this form of familial CRC. On the basis of allele-sharing probabilities, it was estimated that the 9q locus accounted for ∼36% of the inherited susceptibility to CRC. Some support for linkage to this locus was initially provided by two other studies, including our own, but these were not independently statistically significant. Loci mapping to chromosomes 11 and 14 have also been proposed as the position of novel CRC predisposition loci in an analysis of 18 CRC families on the basis of metrics suggestive of linkage, neither loci being statistically robust.17 Recently, Wiesner and co-workers18 reported a GWLS based on a combined analysis of their original families together with 141 kindreds, affection being defined by multiple cancer types including breast cancer and CRA, but including at least one family member with early onset CRC or CRA. On the basis of the analysis, four additional CRC loci at 1p31.1, 15q14-q22, 17p13.3 and 21 were proposed. The observation of linkage, albeit nonsignificant, is interesting as this region includes the HMPS/CRAC1 locus associated with HMPS syndrome in families of Ashkenazi descent.19
In 2006, we reported findings from a GWLS of 69 families with HNSCRC and this provided evidence for a novel susceptibility locus on 3q21–q24. To further evaluate this locus and search for linkage at other loci, we conducted an additional GWLS of a further 34 HNSCRC families. As before, we made use of high-density SNP arrays to search for CRC predisposition loci by linkage analysis. In addition to affording maximal power to detect linkage, the output from such arrays permits efficient pooling of data from different scans, avoiding many of the serious problems associated with microsatellite-based searches.
In our combined analysis, there was no evidence of linkage to the previously proposed loci at 1p, 9q, 11, 15q, 14, 17q or 21. It is possible that failure to demonstrate linkage to these loci is a consequence of differences in the disease phenotype or affection statuses in families analysed. Unusually, the linkage analyses conducted by Wiesner and co-workers6, 18 made use of data from both discordant and concordant sibling pairs. On the basis of the purported contribution of the 9q locus to the familial risk of CRC and the difficulty in assigning ‘unaffected’ status on the basis of presence or absence of adenomas on colonoscopic examination, the robustness of the finding of a disease locus at loci such as 9q22.2 is questionable.
On the basis of an analysis of families in which we restricted affection status to a diagnosis of CRC, rather than using more general criteria including adenomas, maximal linkage was attained at 3q22 in our combined data set under a dominant model. The majority of evidence did, however, come from the original set of families analysed. This is likely in part to be a consequence of the high proportion of the families analysed in phase 2 comprising affected sibships, favouring recovery of a recessive model.
By combining data from the two GWLSs, we have been able to refine our estimate of the impact of the 3q locus on familial CRC. The risk of CRC in young relatives of early-onset CRC cases is increased approximately 10-fold.1 On the basis of allele-sharing probabilities, the 3q22 locus is likely to account for approximately 30% of the familial risk.
All of the patients with adenomas whom we classed as affected had unusually severe disease in terms of tumour multiplicity, age of onset or histology. Our estimates of phenocopy rates for these cases were inevitably imprecise, but were likely to be very conservative. Indeed, almost identical criteria to ours have recently been found independently to be a predictor of adenoma recurrence and hence probably of disease predisposition. We therefore expect all of our affected patients who have adenomas but not CRC to be at greatly increased risk of the latter. Nevertheless, different loci may favour adenoma occurrence and progression to carcinoma, as illustrated by comparing the Mendelian conditions of FAP and hereditary non-polyposis colon cancer. Our finding of significant linkage based on CRC affection status in a subset of families is not, therefore, unexpected. Accepting the caveat that we had only incomplete pathological data on tumour site, it is of interest that those families displaying most evidence for linkage had a high frequency of rectal disease, raising the possibility of some site specificity with respect to disease risk.
In addition to the evidence of linkage of CRC to chromosome 3q22 in the combined analysis, we found suggestive evidence of linkage in the whole data set to chromosome 18q21 and chromosome 2p22, the former based on the NPL statistic and the latter on a recessive model of inheritance. It is possible that there may be epistatic interactions between these putative loci, but data from the current analysis are too limited to address such a postulate.
Forty-nine known genes map to the 8 Mb region of maximal linkage at 3q21–22. Despite an exhaustive mutational analysis of 30 genes prioritized on the basis of candidacy and position of maximal linkage, we were unable to identify a causal variant. Although the finding of a truncating mutation in A4GNT segregating in one family was intriguing, the mutation was demonstrable in the general population. Furthermore, the fact that A4GNT is not expressed in the colon mucosa20 makes this gene a highly unattractive candidate for the 3q22 linkage.
Our analysis was, however, confined to screening of exons and splice sites, and it is possible that causal variants localize to promoter or other regulatory regions. Alternatively, causal variants may reside within one of the 20 unscreened genes or three hypothetical transcripts annotated within 3q21–22 in NCBI Build 36.1 (Mar, 2006; hg18). Further analyses are required to address this possibility, such as high-throughput resequencing technologies. On the basis of our LD mapping studies, it is highly unlikely that a common genetic variant is responsible for the observed linkage, but it remains possible that rare high- or moderate-penetrance alleles outside the genes screened contribute to CRC risk.
References
Johns LE, Houlston RS : A systematic review and meta-analysis of familial colorectal cancer risk. Am J Gastroenterol 2001; 96: 2992–3003.
Aaltonen L, Johns L, Jarvinen H, Mecklin JP, Houlston R : Explaining the familial colorectal cancer risk associated with mismatch repair (MMR)-deficient and MMR-stable tumors. Clin Cancer Res 2007; 13: 356–361.
Bonaiti-Pellie C : Genetic risk factors in colorectal cancer. Eur J Cancer Prev 1999; 8 (Suppl 1): S27–S32.
Peel DJ, Ziogas A, Fox EA et al: Characterization of hereditary nonpolyposis colorectal cancer families from a population-based series of cases. J Natl Cancer Inst 2000; 92: 1517–1522.
Kemp Z, Carvajal-Carmona L, Spain S et al: Evidence for a colorectal cancer susceptibility locus on chromosome 3q21–q24 from a high-density SNP genome-wide linkage scan. Hum Mol Genet 2006; 15: 2903–2910.
Wiesner GL, Daley D, Lewis S et al: A subset of familial colorectal neoplasia kindreds linked to chromosome 9q22.2–31.2. Proc Natl Acad Sci USA 2003; 100: 12961–12965.
Skoglund J, Djureinovic T, Zhou XL et al: Linkage analysis in a large Swedish family supports the presence of a susceptibility locus for adenoma and colorectal cancer on chromosome 9q22.32–31.1. J Med Genet 2006; 43: e7.
Kemp ZE, Carvajal-Carmona LG, Barclay E et al: Evidence of linkage to chromosome 9q22.33 in colorectal cancer kindreds from the United Kingdom. Cancer Res 2006; 66: 5003–5006.
Abecasis GR, Cherny SS, Cookson WO, Cardon LR : Merlin – rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 2002; 30: 97–101.
Boyles AL, Scott WK, Martin ER et al: Linkage disequilibrium inflates type I error rates in multipoint linkage analysis when parental genotypes are missing. Hum Hered 2005; 59: 220–227.
Webb EL, Sellick GS, Houlston RS : SNPLINK: multipoint linkage analysis of densely distributed SNP data incorporating automated linkage disequilibrium removal. Bioinformatics 2005; 21: 3060–3061.
Gudbjartsson DF, Jonasson K, Frigge ML, Kong A : Allegro, a new computer program for multipoint linkage analysis. Nat Genet 2000; 25: 12–13.
Chen CD, Yen MF, Wang WM, Wong JM, Chen TH : A case–cohort study for the disease natural history of adenoma-carcinoma and de novo carcinoma and surveillance of colon and rectum after polypectomy: implication for efficacy of colonoscopy. Br J Cancer 2003; 88: 1866–1873.
Faraway JJ : Distribution of the admixture test for the detection of linkage under heterogeneity. Genet Epidemiol 1993; 10: 75–83.
Risch N : Linkage strategies for genetically complex traits. I. Multilocus models. Am J Hum Genet 1990; 46: 222–228.
Adie EA, Adams RR, Evans KL, Porteous DJ, Pickard BS : SUSPECTS: enabling fast and effective prioritization of positional candidates. Bioinformatics 2006; 22: 773–774.
Djureinovic T, Skoglund J, Vandrovcova J et al: A genome wide linkage analysis in Swedish families with hereditary non-familial adenomatous polyposis/non-hereditary non-polyposis colorectal cancer. Gut 2006; 55: 362–366.
Daley D, Lewis S, Platzer P et al: Identification of susceptibility genes for cancer in a genome-wide scan: results from the colon neoplasia sibling study. Am J Hum Genet 2008; 82: 723–736.
Jaeger EE, Woodford-Richens KL, Lockett M et al: An ancestral Ashkenazi haplotype at the HMPS/CRAC1 locus on 15q13–q14 is associated with hereditary mixed polyposis syndrome. Am J Hum Genet 2003; 72: 1261–1267.
Nakajima K, Ota H, Zhang MX et al: Expression of gastric gland mucous cell-type mucin in normal and neoplastic human tissues. J Histochem Cytochem 2003; 51: 1689–1698.
Acknowledgements
We are grateful to the patients, their families and their clinicians for their participation in this study. The CORGI Consortium comprised Lucy Side, Lucy Curtis and Peter Risby (Oxford Regional Genetics Service, Churchill Hospital, Oxford, UK), Carole Cummings and Angela Brady (North-West Thames Regional Genetics Service, Northwick Park Hospital, Harrow, UK), Joan Paterson (Anglia Regional Genetics Service, Addenbrooke's Hospital, Cambridge, UK), Shirley Hodgson and Tessa Homfray (South-West Thames Regional Genetics Service, St George's Hospital, Tooting, London, UK), Louise Izatt (South-East Thames Regional Genetics Service, Guy's Hospital, London, UK), Alan Donaldson (South-West Regional Genetics Service, Bristol, UK), Patrick Morrison (Northern Ireland Regional Genetics Service, City Hospital, Belfast, UK), Carole Brewer (South-West Regional Genetics Service, Royal Devon and Exeter Hospital, Exeter, UK), John Burn and Alison Trainer (Northern Regional Genetics Service, International Centre for Life, Newcastle, UK), Rosemarie Davidson and Victoria Murday (West of Scotland Regional Genetics Service, Yorkhill Hospital, Glasgow, UK), Jaqueline Cook (Sheffield Regional Genetics Service, Children's Hospital, Sheffield, UK), Neva Haites (North of Scotland Regional Genetics Service, Foresterhill Hospital, Aberdeen, UK), Eamonn Sheridan (Yorkshire Regional Genetics Service, St James's Hospital, Leeds, UK), Andrew Green (Republic of Ireland Genetics Service, Our Lady's Hospital for Sick Children, Dublin, Republic of Ireland) and Susan Ritchie (Nottingham Centre for Medical Genetics, City Hospital, Nottingham, UK), who should be regarded as co-authors of this work. The work was supported by grants from Cancer Research UK. ZK was funded by the EU, GS by Leukemia Research, EP by the Institute of Cancer Research and SS by CORE. Finally, work of the German HNPCC consortium is supported by German Cancer Aid.
Author information
Authors and Affiliations
Consortia
Corresponding authors
Additional information
Web addrresses
URLs for programs and data presented herein are as follows:
Online Mendelian Inheritance in Man (OMIM): http://www.ncbi.nlm.nih.gov/sites/entrez
SUSPECTS: http://www.genetics.med.ed.ac.uk/suspects/
dbSNP: http://www.ncbi.nlm.nih.gov/sites/entrez
UCSC Human Genome browser, http://genome.ucsc.edu/cgi-bin/hgGateway
Affymetrix NetAffx: http://www.affymetrix.com/analysis/index.affx
Illumina: http://www.illumina.com/
SNPLINK: http://www.icr.ac.uk/cancgen/molgen/MolPopGen_Bioinformatics.htm
R suite of programs: http://www.r-project.org/
Conflict of interest
The authors state no conflict of interest.
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Rights and permissions
About this article
Cite this article
Papaemmanuil, E., Carvajal-Carmona, L., Sellick, G. et al. Deciphering the genetics of hereditary non-syndromic colorectal cancer. Eur J Hum Genet 16, 1477–1486 (2008). https://doi.org/10.1038/ejhg.2008.129
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2008.129
Keywords
This article is cited by
-
Molecular Analysis of Iranian Colorectal Cancer Patients at Risk for Lynch Syndrome: a New Molecular, Clinicopathological Feature
Journal of Gastrointestinal Cancer (2015)
-
Genome-wide linkage analysis and tumoral characterization reveal heterogeneity in familial colorectal cancer type X
Journal of Gastroenterology (2015)
-
Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas
Nature Genetics (2013)
-
Evidence of linkage to chromosomes 10p15.3–p15.1, 14q24.3–q31.1 and 9q33.3–q34.3 in non-syndromic colorectal cancer families
European Journal of Human Genetics (2012)
-
Hereditary mixed polyposis syndrome is caused by a 40-kb upstream duplication that leads to increased and ectopic expression of the BMP antagonist GREM1
Nature Genetics (2012)