Abstract
Experimental genetics have been widely used to explore the biology of the malaria parasites. The rodent parasites Plasmodium berghei and less frequently P. yoelii are commonly utilised, as their complete life cycle can be reproduced in the laboratory and because they are genetically tractable via homologous recombination. However, due to the limited number of drug-selectable markers, multiple modifications of the parasite genome are difficult to achieve and require large numbers of mice. Here we describe a novel strategy that combines positive-negative drug selection and flow cytometry-assisted sorting of fluorescent parasites for the rapid generation of drug-selectable marker-free P. berghei and P. yoelii mutant parasites expressing a GFP or a GFP-luciferase cassette, using minimal numbers of mice. We further illustrate how this new strategy facilitates phenotypic analysis of genetically modified parasites by fluorescence and bioluminescence imaging of P. berghei mutants arrested during liver stage development.
Similar content being viewed by others
Introduction
Malaria is caused by single cell eukaryotes belonging to the genus Plasmodium, the deadliest of which is Plasmodium falciparum. Rodent malaria parasites such as P. berghei and P. yoelii provide useful models to study malaria as their life cycles can be completed in the laboratory by cycling between infected mice and Anopheles stephensi mosquitoes. Furthermore, these parasites are genetically tractable. Experimental genetics have been extensively utilised to gain an in-depth understanding of the biology of the parasites and interactions with their hosts1. Targeted gene deletion and protein tagging can provide insights into the function of Plasmodium genes and the proteins they encode. In addition, experimental genetics have also been exploited to generate parasites expressing heterologous transgenes, such as the green fluorescent protein (GFP) or bioluminescent luciferase (LUC) reporter probes to visualise and analyse parasite-host interactions in vitro and in vivo2.
Genetic manipulation of the parasites relies on homologous recombination between a target sequence in the parasite genome and a DNA construct that contains a selection cassette3. Blood stage parasites are transfected with a DNA construct and the recombinant parasites are allowed to replicate asexually prior to selection4. Standard protocols to generate transgenic rodent malaria parasites usually require high numbers of animals at multiple steps, including cloning of the recombinant parasite population by limiting dilutions and injection into mice. Recently, flow cytometry-assisted sorting of GFP-expressing transgenic parasites has been used to isolate isogenic populations of P. berghei mutants, without the need for in vivo cloning5,6. Only two drug-selectable markers – a modified form of the dihydrofolate reductase-thymidylate synthase (DHFR-TS) from Toxoplasmagondii or P. berghei and the human DHFR (hDHFR) - can be used to generate transgenic rodent malaria parasites. These markers confer resistance to pyrimethamine (in the case of Tg or PbDHFR-TS) or both pyrimethamine and WR99210 (in the case of hDHFR) and can only be used sequentially7, thus severely limiting the range of genetic manipulations that can be made in one single parasite. Recently, a positive-negative selection procedure has been developed to recycle the selectable marker and allow successive modifications of the parasite genome8,9. This procedure utilises the bifunctional yeast enzyme cytosine deaminase and uridyl phosphoribosyl transferase (yFCU), as a negative selectable marker. The yFCU enzyme converts the pro-drug 5-fluorocytosine (5-FC) into the highly toxic compound 5-fluorouracil (5-FU). Initially, transformed parasites expressing a hDHFR-yFCU fusion gene are positively selected with pyrimethamine. Subsequent negative selection is performed with 5-FC, which is converted into toxic 5-FU in parasites expressing yFCU, resulting in the death of parasites harbouring the hDHFR-yFCU marker. This allows the recovery of marker-free parasites that have excised the hDHFR-yFCU marker after homologous recombination around the selection cassette. However, negative selection with 5-FC does not eliminate 100% of marker-containing parasites, therefore isolation of pure populations of marker-free parasites requires an additional cloning step performed in vivo in mice8,9,10,11.
We have now combined positive-negative selection and flow cytometry-assisted sorting of fluorescent recombinant parasites into a single procedure, termed GOMO (‘Gene Out Marker Out’). This new selection method totally eliminates the need for in vivo cloning of the parasites and allows the rapid generation of drug-selectable marker-free P. berghei and P. yoelii mutants expressing a GFP or GFP-LUC cassette, using as few as three mice. This strategy will facilitate genetic screens in rodent malaria parasites, allowing combined genetic modifications and phenotypic analysis of genetically modified parasites by fluorescence or bioluminescence imaging.
Results
The GOMO strategy: ‘Gene Out, Marker Out’
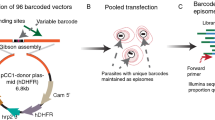
Our goal was to integrate recent advances in experimental genetics protocols into one single novel strategy, termed GOMO, for the rapid isolation of pure populations of recombinant rodent malaria parasites. This strategy allows concomitant replacement of a gene of interest by a fluorescent or luminescent cassette (‘Gene Out’) and removal of the drug-selectable marker (‘Marker Out’), thus facilitating both downstream phenotypic analysis and further genetic modifications (Fig. 1). We first assembled two different GOMO plasmids, containing a GFP or a GFP-LUC cassette under control of the constitutive promoter of PbHSP70 (PBANKA_071190) or PbeEF1α (PBANKA_113330), respectively (Fig. S1). The HSP70 promoter allows strong and constitutive expression of GFP at all stages of parasite development, including sporozoites and is thus ideal for imaging purposes12. Attempts to use the HSP70 promoter to express GFP-LUC failed, possibly due to deleterious effects of the fusion protein on parasite growth. Therefore we used the eEF1α promoter to drive constitutive expression of GFP-LUC and allow live imaging of the parasite in vivo by bioluminescence, including in the liver13,14. In addition to the GFP (or GFP-LUC) cassette, both GOMO plasmids contain a hDHFR-yFCU fusion gene, for positive-negative selection8, coupled to a second fluorescent cassette, encoding the red fluorescent protein mCherry (Fig. S1). Both hDHFR-yFCU and mCherry are placed under control of a single bidirectional promoter of PbeEF1α (PBANKA_113330 and PBANKA_113340). The GFP (or GFP-LUC) and mCherry reporter genes are followed by an identical 1 kb fragment corresponding to the 3′ untranslated region (UTR) of P. berghei DHFR-TS, placed in the same orientation, which serves both as a transcription terminator, for proper expression of the reporters and for recycling of the drug resistance cassette. Spontaneous recombination between the two homologous PbDHFR-TS 3′ UTR fragments results in excision of both the hDHFR-yFCU and the mCherry expression cassettes (Fig. 1). Therefore, with this strategy, parasites that have excised the drug-selectable marker become GFP+ mCherry− and can be easily distinguished from GFP+ mCherry+ parasites still harbouring the hDHFR-yFCU marker.
The GOMO strategy: ‘Gene Out Marker Out’.
(A–B). Constructs for targeted gene deletion are assembled by cloning a 5′ and a 3′ homology sequences of the target gene on each side of a triple cassette, consisting of a GFP (A) or GFP-LUC (B) cassette (green box) under control of the PbHSP70 (A) or the PbeEF1α (B) promoter, respectively, a hDHFR-yFCU cassette (blue box) and a mCherry cassette (red box). The hDHFR-yFCU and mCherry genes are both under control of a single bidirectional PbeEF1α promoter. Upon a double crossover recombination event, the target gene is replaced by the GFP(-LUC)/hDHFR-yFCU/mCherry triple cassette. A second recombination event between the two identical PbDHFR/TS 3′ UTR sequences (pink lollipops) results in excision of the hDHFR-yFCU and mCherry cassettes. (C). Overview of the selection procedure. After transfection, parasites are injected into a first mouse, followed by positive selection of recombinant parasites with pyrimethamine. GFP+ mCherry+ pyrimethamine-resistant parasites are then recovered and transferred into a second mouse and exposed to 5-FC for negative selection of parasites that have not excised the hDHFR-yFCU marker. GFP+ mCherry− 5-FC-resistant parasites, which harbour the intended recombined locus after marker excision, are then sorted by FACS and injected into a third mouse, allowing the isolation of a pure population of drug-selectable marker-free recombinant parasites.
The GOMO plasmids can be used for targeted gene deletion, after introduction of 5′ and 3′ homologous sequences from a target gene on each side of the selection cassettes (Fig. 1A and 1B). After transfection of blood stage parasites, a double crossover homologous recombination event results in replacement of the target gene by the GOMO construct. The selection procedure requires as few as three mice and is described in Fig. 1C. Transfected parasites are injected in the first mouse. Positive selection of parasites having incorporated the construct is performed by exposure to pyrimethamine. GFP+ mCherry+ pyrimethamine-resistant parasites, resulting from integration of the construct in the parasite genome or its persistence as an episome, are then recovered and transferred to the second mouse. Parasites are then exposed to 5-FC, which kills parasites containing the yFCU gene. This negative selection step allows the recovery of 5-FC-resistant parasites that have either lost the episome and are therefore GFP− mCherry−, or have recombined the integrated GOMO construct at the homologous PbDHFR-TS 3′ UTR sequences and are therefore GFP+ mCherry− (Fig. 1A and 1B). GFP+ mCherry− blood stage parasites are then sorted by FACS (Fig. S2) and injected into the third mouse, for the isolation of a pure population of recombinant parasites (Fig. 1C). Thus, negative selection is used both to isolate parasites with the intended genome modification and at the same time recycle the drug-selectable marker. Both the positive and negative selection steps take ~1 week, so the whole procedure allows the generation of final mutant parasites in ~3 weeks.
Generation of drug-selectable marker-free Δp230p-GFP and Δp230p-GFP-LUC P. berghei ANKA parasites
To validate the GOMO strategy, we first targeted the dispensable P. bergheiP230p gene (PBANKA_030600). P230p-lacking parasites have been shown to have no phenotypical defect at any stage and progress normally through the whole parasite life cycle15. For this reason, the P230p locus has been chosen in the past to generate reference P. berghei ANKA parasite lines constitutively expressing GFP or GFP-LUC5,10. We generated two PbP230p targeting vectors by inserting a 5′ and a 3′ homology fragment in both GOMO-GFP and GOMO-GFP-LUC plasmids (Fig. 2A and 2B). Wild type (WT) P. berghei blood stage parasites were transfected with linearized constructs and positive selection with pyrimethamine was performed in vivo in mice. FACS analysis of pyrimethamine-resistant parasites revealed the presence of an expected GFP+ mCherry+ parasite population (Fig. 2C and 2D, left panels). The intensity of the GFP fluorescence was higher in parasites that were transfected with the GOMO-GFP vector (Fig. 2C) than in those transfected with the GOMO-GFP-LUC vector (Fig. 2D). This is likely due to the stronger activity of the HSP70 promoter6,16 and possibly also to the lower activity of GFP when fused to LUC. Genotyping by Southern blot (Fig. 2E) and PCR (Fig. 2F and 2G, left panels) confirmed integration of the construct at the PbP230p locus. Parasites with a WT PbP230p locus were still detected at this stage by PCR, indicating that a fraction of the pyrimethamine-resistant parasites retained the construct either as an episome or integrated elsewhere than at the P230p locus (see below). Pyrimethamine-resistant GFP+ mCherry+ parasites were then sorted by FACS (Fig. S2A), injected into new mice and exposed to 5-FC for negative selection and subsequent recovery of 5-FC-resistant parasites. As expected, FACS analysis of 5-FC-resistant parasites revealed the presence of a population of GFP+ mCherry− parasites (Fig. 2C and 2D, right panels). Moreover, genotyping by PCR confirmed that excision of the drug-selectable cassette had occurred and revealed the persistence at this stage of parasites with a WT P230p locus (Fig. 2F and 2G, middle panels). Also, a residual population of GFP+ mCherry+ parasites was still detectable by FACS after 5-FC negative selection, showing that 5-FC treatment is not 100% effective, as reported previously8,10,11.
Generation of drug-selectable marker-free Δp230p-GFP and Δp230p-GFP-LUC P. berghei ANKA parasites.
(A–B). Replacement strategy to generate Δp230p-GFP (A) and Δp230p-GFP-LUC (B) parasites. The wild-type (WT) genomic locus of P. bergheiP230p was targeted with GOMO-GFP (A) and GOMO-GFP-LUC (B) replacement plasmids containing a 5′ and a 3′ homologous sequence inserted on each side of the plasmid cassettes. Upon a double crossover event, the P230p gene is replaced by the GFP(-LUC)/hDHFR-yFCU/mCherry triple cassette. Recombination between the two identical PbDHFR/TS 3′ UTR sequences (pink lollipops) results in excision of hDHFR-yFCU and mCherry. Genotyping primers and expected PCR fragments are indicated by arrows and lines, respectively. The Southern probe and expected restriction fragments are also shown. (C–D). FACS scatter plots of parasites transfected with GOMO-GFP (C) and GOMO-GFP-LUC (D) constructs targeting PbP230p. GFP+ mCherry+ parasites obtained after positive selection with pyrimethamine (left panels) are replaced by GFP+ mCherry− parasites after negative selection with 5-FC (right panels). (E). Southern blot analysis of genomic DNA isolated from P. berghei WT, Δp230p-GFP and Δp230p-GFP-LUC parasites after positive selection with pyrimethamine (1st) or negative selection by 5-FC followed by FACS sorting of GFP+ mCherry− parasites (final), using a probe specific for the 3′ homology sequence of PbP230p. After digest with NheI and AflII, the probe hybridizes to fragments of 8.3 kb in WT, 5.5 kb in recombined non-excised parasites and 5.0 or 7.3 kb after marker excision in the final Δp230p-GFP and Δp230p-GFP-LUC parasite populations, respectively. (F–G). PCR analysis of genomic DNA isolated from P. berghei WT, Δp230p-GFP (F) and Δp230p-GFP-LUC (G) parasites recovered after the first positive selection with pyrimethamine (left panels), after the second negative selection with 5-FC (middle panels) and after flow cytometry sorting of GFP+ mCherry− parasites (final populations, right panels). Confirmation of the predicted recombination events was achieved with primer combinations specific for 5′ or 3′ integration. An additional primer combination was used to assess removal of the hDHFR-yFCU selectable marker (3′ excised). A wild type-specific PCR reaction (WT) confirmed the absence of residual wild-type parasites in the final GFP+ mCherry− FACS-sorted parasites.
To isolate pure recombinant populations, GFP+ mCherry− parasites were sorted by flow cytometry, using the gating strategy shown in Fig. S2B and injected into a naive mouse. Genotyping of the resulting parasite populations by Southern blot (Fig. 2E) and PCR (Fig. 2F and 2G, right panels) showed that all parasites had a deleted P230p locus and had excised the drug-selectable marker and mCherry cassettes. In summary, using the GOMO strategy, we could successfully generate pure polyclonal populations of drug-selectable marker-free Δp230p-GFP and Δp230p-GFP-LUC P. berghei ANKA parasites, using less than 5 mice each and without any in vivo cloning step.
Alternative recombination events are efficiently discriminated by the GOMO procedure
FACS analysis after the initial positive selection step revealed the presence of a GFP− mCherry+ population among pyrimethamine-resistant parasites (Fig. 2C and 2D, left panels). We hypothesized that these parasites might result from alternative integration of the GOMO constructs at the P.berghei DHFR-TS locus, due to the presence of the two homologous PbDHFR-TS 3′ UTR fragments flanking the hDHFR-yFCU and mCherry cassettes (Fig. S3A and S3B). The occurrence of a double crossover at the parasite DHFR-TS locus was confirmed by PCR genotyping of pyrimethamine-resistant parasites recovered after transfection with both GOMO-GFP (Fig. S3C) and GOMO-GFP-LUC (Fig. S3D) PbP230p targeting vectors. Nevertheless, it should be noted that undesired integration of the constructs at the DHFR-TS locus is eliminated after negative selection by 5-FC, as recombination and excision of the hDHFR-yFCU cassette results in restoration of a WT locus and absence of fluorescence of the resulting parasites (Fig. S3A and S3B). Furthermore, flow cytometry sorting of GFP+ mCherry+ parasites after positive selection (Fig. 1C) eliminates parasites that have integrated the construct at the DHFR-TS locus. Theoretically, the GOMO-GFP-LUC construct could also integrate at the eEF1α locus, due to the presence of two copies of the eEF1α (PBANKA_113330) promoter (Fig. S4). However, we were unable to detect such recombination event in any of the transfection experiments performed in this study (data not shown), perhaps because double crossover recombination in the short repeated sequence (586-bp) is a rare event, or because the eEF1α locus is refractory to genetic modification. In any case, such undesired integration of the constructs at the eEF1α locus would be eliminated after negative selection by 5-FC, as recombination and excision of the hDHFR-yFCU cassette results in the restoration of a WT locus (Fig. S4).
Interestingly, after 5-FC negative selection of GOMO-GFP-LUC transfected parasites, we still observed a GFP− mCherry+ population in addition to the expected GFP+ mCherry− population (Fig. 2D, right panel). Furthermore, genotyping by PCR revealed the persistence of parasites with a non-excised 3′ recombined P230p locus (Fig. 2G, middle panel). These 5-FC-resistant GFP− mCherry+ parasites were sorted by FACS and genotyped by PCR and DNA sequencing. The results indicate that this population corresponds to Δp230p mutants that have excised the GFP-LUC and hDHFR-yFCU cassettes, after recombination in the two identical eEF1α promoter sequences contained in the construct (Fig. S5). Therefore, in addition to Δp230p-GFP-LUC mutants, the GOMO strategy allowed us to isolate a pure population of drug-selectable marker-free Δp230p-mCherry P. berghei ANKA parasites.
Generation of marker-free Δslarp-GFP and Δslarp-GFP-LUC P. berghei ANKA parasites
To illustrate the usefulness of the approach for phenotypic analyses, we used the GOMO strategy to generate GFP- and GFP-LUC-expressing P. berghei mutants lacking the Sporozoite and Liver Stage Asparagin-Rich Protein (SLARP, PBANKA_090210). SLARP is a conserved Plasmodium gene that is specifically expressed at pre-erythrocytic stages and is required for parasite development in the liver17,18. SLARP-deficient sporozoites injected into susceptible mice cannot convert into blood stages in vivo. They invade cells efficiently in vitro, but then rapidly disappear from cultures in the first 24 hours17,18. Two PbSLARP targeting vectors were assembled by inserting 5′ and 3′ SLARP gene homology fragments in GOMO-GFP and GOMO-GFP-LUC plasmids (Fig. S6A and S6B) and used to transfect WT P. berghei ANKA blood stage parasites. We then applied the GOMO selection strategy, as described above, to isolate pure Δslarp-GFP and Δslarp-GFP-LUC parasite lines. As expected, FACS analysis revealed the presence of a GFP+ mCherry+ parasite population after pyrimethamine treatment (Fig. 3A and 3B, left panels). After negative selection with 5-FC, we obtained GFP+ mCherry− parasites (Fig. 3A and 3B, right panels), which were then isolated by flow cytometry-assisted sorting. Genotyping of the resulting parasite populations by PCR (Fig. 3C) and Southern blot (Fig. 3D) confirmed deletion of SLARP gene and excision of the drug-selectable marker and the mCherry cassette, as desired. Pure populations of Δp230p-GFP, Δp230p-GFP-LUC, Δslarp-GFP and Δslarp-GFP-LUC were then transmitted to A. stephensi mosquitoes in order to obtain sporozoites.
Generation of drug-selectable marker-free Δslarp-GFP and Δslarp-GFP-LUC P. berghei ANKA parasites.
(A–B). FACS scatter plots of parasites transfected with GOMO-GFP (A) and GOMO-GFP-LUC (B) constructs targeting PbSLARP (PBANKA_090210). The GFP+ mCherry+ parasite population obtained after positive selection with pyrimethamine (left panels) was replaced by a GFP+ mCherry− population after negative selection with 5-FC (right panels). (C). PCR analysis of genomic DNA isolated from P. berghei WT, Δslarp-GFP (left panel) and Δslarp-GFP-LUC (right panel) parasites recovered after GOMO selection. Confirmation of the predicted recombination events was achieved with primer combinations specific for 5′ integration (5′ integr.) or 3′ integration followed by marker excision (3′ excised). Primers used for genotyping are indicated in Fig. S6 and Table S1. The absence of amplification with primer combinations specific for the WT locus (WT) and the non-excised integrated construct (3′ integration) confirmed that the final populations contained only Δslarp marker-free parasites. (D). Southern blot analysis of genomic DNA isolated from P. berghei WT, Δslarp-GFP and Δslarp-GFP-LUC final parasite populations selected with the GOMO procedure, using a digoxigenin-labelled probe specific for the 3′ homology sequence of PbSLARP. After digest with SnaBI and AflII, the probe hybridizes to a 11.6 kb fragment in WT, a 4.0 kb fragment in recombined non-excised parasites and a 3.6 or 5.8 kb fragment after marker excision in the final Δslarp-GFP and Δslarp-GFP-LUC parasite populations, respectively. The position of the probe and the restriction fragments are indicated in Fig. S6.
We then analysed the phenotype of the Δslarp-GFP-LUC and Δslarp-GFP P. berghei mutants, using Δp230p-GFP-LUC and Δp230p-GFP parasites as controls. We first used intravital imaging of bioluminescent parasites to analyse the fate of Δslarp P. berghei sporozoites in vivo after intravenous inoculation into C57BL/6 mice. Control Δp230p-GFP-LUC parasites were readily detectable in the liver of infected mice 24 h after injection of sporozoites (Fig. 4A and 4D) and the signal further increased at 48 h (Fig. 4B and 4E). All mice developed a patent parasitemia, as shown by positive Giemsa-stained blood smears and diffusion of the luminescence signal in the entire mouse body at day 4 post-infection (Fig. 4C and 4F). In sharp contrast, mice injected with Δslarp-GFP-LUC showed no luminescence signal above background at any of the time points examined (Fig. 4A–F) and none of the animals became patent, in total agreement with published data showing that ΔslarpP. berghei sporozoites fail to convert into blood stages18.
Intravital bioluminescence imaging of Δp230p-GFP-LUC and Δslarp-GFP-LUC P. berghei liver stage infection.
(A–C). C57BL/6 mice were injected intravenously with 50,000 sporozoites of control Δp230p-GFP-LUC (left panels) or mutant Δslarp-GFP-LUC (right panels) and imaged at 24, 48 and 96 hours post-infection after intraperitoneal injection of D-luciferin. (E–F). Quantification of luminescence levels in the whole body of mice infected with Δp230p-GFP-LUC (grey bars) or mutant Δslarp-GFP-LUC (black bars). Data are expressed as photons/second (mean +/− sd, **p < 0.01, Mann-Whitney test). Dotted red lines represent baseline measurement of naive mice.
To analyse in more details the fate of Δslarp sporozoites, we used time-lapse fluorescence microscopy to image control Δp230p-GFP and Δslarp-GFP parasites in HepG2 cultures from 7 to 24 hours post-infection (Fig. 5 and supplemental movies 1–5). Transformation of elongated sporozoites into round forms was observed with both Δp230p-GFP and Δslarp-GFP. However, whilst Δp230p-GFP persisted in the culture and started growing over the observation period (Fig. 5A, 5C and supplemental movies 1–2), Δslarp-GFP failed to develop and died, as evidence by disappearance of the GFP signal from infected cells (Fig. 5B, 5C and supplemental movies 3–5). We observed no detectable sign of death of infected host cells by microscopy, suggesting that elimination of the Δslarp mutant parasites, unlike other liver stage-arresting mutants19, does not result from host cell apoptosis. Altogether, these data illustrate the usefulness of the GOMO approach for generating GFP- or GFP-LUC-expressing mutants and monitoring liver infection in vitro and in vivo by fluorescence microscopy and bioluminescence imaging, respectively.
Time lapse imaging of Δp230p-GFP and Δslarp-GFP P. berghei liver stage infection in vitro.
(A). Time lapse microscopy of a Δp230p-GFP P. berghei sporozoite (green) inside a HepG2 cell, showing parasite transformation and growth. The images were extracted from the supplementary movie 1. The time after sporozoite inoculation (h:min) is indicated in the upper left corner of each image. Bars, 10 μm. (B). Time lapse microscopy of a Δslarp-GFP P. berghei sporozoite (green) inside a HepG2 cell, showing parasite elimination from infected host cells. The images were extracted from the supplementary movie 3. The time after sporozoite inoculation (h:min) is indicated in the upper left corner of each image. Bars, 10 μm. (C). Survival of Δp230p-GFP (n = 22) and Δslarp-GFP (n = 29) intracellular parasites over the imaging period (from 7:25 to 24:45 post-infection) was analysed from a total of 9 movies (p = 0.0003, Mantel-Cox log-rank test).
Generation of marker-free Δp230p-GFP and Δp230p-GFP-LUC P. yoelii 17XNL parasites
Finally, we tested whether the GOMO strategy can be used in the rodent malaria parasite P. yoelii, which has proven to be a relevant model to study different aspects of the malaria infection, such as sporozoite invasion or liver stage immunity20,21. We chose PyP230p (PY03857) as a target gene, as it was recently shown to be dispensable in P. yoelii as well10. Two targeting vectors were assembled by inserting 5′ and 3′ PyP230p gene homology fragments in GOMO-GFP and GOMO-GFP-LUC plasmids (Fig. S7A and S7B) and used to transfect WT P. yoelii 17XNL blood stage parasites. We then applied the same GOMO selection strategy, as described above. FACS analysis revealed the presence of a GFP+ mCherry+ parasite population after pyrimethamine treatment, as expected (Fig. 6A and 6B, left panels). After negative selection with 5-FC, we obtained GFP+ mCherry− parasites (Figure 6A and 6B, right panels), which were isolated by FACS. Genotyping by PCR confirmed deletion of the PyP230p gene and excision of the drug-selectable marker and the mCherry cassette, as desired (Figure 6C). These data demonstrate that GOMO is a robust and efficient strategy for targeted gene deletion not only in P. berghei but also in P. yoelii.
Generation of drug-selectable marker-free Δp230p-GFP and Δp230p-GFP-LUC P. yoelii 17XNL parasites.
(A–B). FACS scatter plots of P. yoelii parasites transfected with GOMO-GFP (A) and GOMO-GFP-LUC (B) constructs targeting PyP230p (PY03857). The GFP+ mCherry+ parasite population obtained after positive selection with pyrimethamine (left panels) was replaced by a GFP+ mCherry− population after negative selection with 5-FC (right panels). (C). PCR analysis of genomic DNA isolated from P. yoelii WT, Δp230p-GFP (left panel) and Δp230p-GFP-LUC (right panel) parasites recovered after GOMO selection. Confirmation of the predicted recombination events was achieved with primer combinations specific for 5′ integration (5′ integr.) or 3′ integration followed by marker excision (3′ excised). Primers used for genotyping are indicated in Fig. S7 and Table S1. The absence of amplification with primer combinations specific for the WT locus (WT) and the non-excised integrated construct (3′ integration) confirms that the final populations contain only Δp230p drug-selectable marker-free P. yoelii parasites.
Discussion
Genetically modified malaria parasites have proven to be valuable tools in elucidating Plasmodium biology and dissecting interactions with both the mosquito and vertebrate hosts. The development of highly effective gene targeting technologies has greatly facilitated the generation of transgenic parasites in the rodent parasite P. berghei and to a lesser extent P. yoelii. A major bottleneck with standard protocols to isolate pure recombinant parasite lines is the mandatory in vivo cloning step, which typically consists of a limiting parasite dilution followed by injection into series of at least 10 mice. Furthermore, the paucity of drug-selectable markers that can be used in vivo has severely limited the range of genetic manipulations that can be made in one single parasite. A positive-negative selection method using the hDHFR-yFCU fusion gene and treatment with the pro-drug 5-FC has been developed to recycle the selectable marker8. As negative selection by 5-FC is not 100% effective, a parasite cloning step remains necessary to isolate pure populations of marker-free parasites8,9,10,11. More recently, flow cytometry-assisted sorting of GFP-expressing transgenic parasites has been used to isolate isogenic populations of P. berghei mutants without the need for in vivo cloning5,6.
We have now integrated these advanced methods into a single procedure, termed GOMO (‘Gene Out Marker Out’), for the rapid production of pure drug-selectable marker-free transgenic parasite populations without the need for in vivo cloning. The strategy is based on the use of an improved targeting plasmid vector, which contains two fluorescence cassettes (GFP and mCherry) and a hDHFR-yFCU fusion gene for positive-negative selection. The hDHFR-yFCU and mCherry cassettes are flanked by an identical sequence to allow excision of the two cassettes by spontaneous homologous recombination. Marker-free mutant parasites are obtained within 3 weeks following transfection, after four sequential selection steps: positive selection with pyrimethamine, flow cytometry sorting of GFP+ mCherry+ parasites, negative selection with 5-FC and flow cytometry sorting of GFP+ mCherry− parasites (Fig. 1C). The first flow cytometry sorting step performed immediately after pyrimethamine selection is used to isolate GFP+ mCherry+ parasites, leading to enrichment for parasites with the desired genetic modification. This step eliminates GFP− mCherry+ parasites that have integrated the construct at the DHFR/TS locus, as well as GFP+ mCherry− parasites that have integrated the construct at the eEF1α locus. This initial FACS step might be omitted when only GFP+ mCherry+ parasites are observed following positive selection with pyrimethamine. Importantly, all sorting experiments must be performed at low parasitemia (<1%), to avoid contamination of the sorted populations by multiply infected erythrocytes.
The GOMO procedure offers a number of advantages over previous protocols. It allows the rapid isolation of drug-selectable marker-free mutants, within 3 weeks, without the need for parasite cloning in vivo, thus reducing the number of experimental animals to a minimum of three mice per mutant (or six mice if experiments are duplicated, for example to generate independent transgenic lines or to increase the chances of successful gene targeting). In comparison, currently available protocols would require at least 15 mice to generate pure marker-free parasite lines, including two mice for the initial positive selection and flow cytometry-assisted sorting of GFP-expressing parasites6 and an additional 13–15 mice for negative selection and parasite cloning9,10. Furthermore, at least two independent clones, obtained from independent transfection experiments, are usually required for phenotypic analysis, further increasing animal usage. The GOMO procedure generates genetically homogenous (isogenic) polyclonal populations, with the advantage of minimizing the risk of clone-specific phenotypic defects unrelated to the intended gene modification. Inclusion of a dual fluorescence cassette in the GOMO vectors allows specific isolation of recombined marker-free parasites by FACS, thus eliminating the need for parasite cloning. It also facilitates monitoring of the successive selection steps, allowing discrimination not only of the marker excision event but also of alternative recombination events, such as integration of the construct at the parasite DHFR/TS locus. Another advantage of the GOMO procedure is that the recycling of the marker is achieved in the same transfection experiment, in contrast to the GIMO protocol (‘Gene Insertion/Marker Out’), where a second transfection is necessary to replace the drug-selectable marker10. In addition, with the GOMO-GFP-LUC vector, both GFP-LUC- and mCherry-expressing marker-free parasites can be obtained from the same transfection experiment. The GOMO transfections can be performed in any parasite line and do not require the use of a parasite mother line as with the GIMO protocol10. A similar strategy could be used to isolate transgenic lines in P. falciparum, where positive-negative selection also operates22.
Recycling of the drug-selectable marker greatly facilitates downstream genetic modifications in the same mutant, including additional gene deletion, transgene expression and genetic complementation8,10. GOMO generates GFP or GFP-LUC expressing parasites, thus facilitating downstream phenotypic analysis of mutants by fluorescence and bioluminescence imaging2. Finally, after transfer of the GOMO cassettes into a Gateway-compatible plasmid backbone, the selection procedure could be combined with the PlasmoGEM recombineering system, which has been recently developed for high-throughput, genome wide and highly efficient generation of gene targeting constructs23.
Whilst mice remain necessary for the production of genetically modified rodent malaria parasites, the GOMO method significantly contributes to both reduction and refinement of animal usage. The strategy allows a drastic reduction of the number of mice used to produce mutant lines. Furthermore, the protocol uses refinement methods to perform positive and negative selection via oral administration of pyrimethamine and 5-FC in the drinking water, as recently described9. Finally, introduction of the GFP-LUC transgene allows non-invasive quantification of parasite burden in the same mouse over the course of infection, especially during the liver stage development, further reducing animal usage for phenotypical analysis.
To establish the GOMO selection procedure we generated Δp230p-GFP and Δp230p-GFP-LUC marker-free parasite lines in both P. berghei ANKA and P. yoelii 17XNL. The P230p locus, which is phenotypically silent in both parasite species10,15, has been targeted before to generate the GFP reference P. berghei ANKA 507 clone5 and more recently GFP-LUC marker-free P. berghei and P. yoelii parasites10. We have now produced the first drug-selectable marker-free P. yoelii line constitutively expressing GFP under control of the HSP70 promoter, which allows bright fluorescence at all stages of the parasite life cycle6,16 and is thus optimal for imaging applications12.
To illustrate its usefulness in the context of reverse genetic screens, we applied the GOMO strategy to generate Δslarp-GFP and Δslarp-GFP-LUC P. berghei mutants and provide the first study of a liver stage-arresting mutant both in the living mouse by bioluminescence imaging and in cultured cells by time lapse fluorescence microscopy. Radiation or genetically attenuated sporozoites arresting early during liver stage development rapidly disappear from in vitro cultures18,19,24,25,26. Whilst the mechanisms of parasite elimination are unknown, one study reported that deletion of the P52 gene in P. berghei causes host cell death by apoptosis19. Here we show by video-microscopy that Δslarp-GFP intracellular parasites fail to develop and survive inside infected cells and rapidly disappear without any sign of host cell death. SLARP is a major regulator of gene expression in sporozoites and liver stages17,18. We have now generated ΔslarpP. berghei lines that are devoid of drug-selectable marker. These parasites can thus be easily modified further, for example to express reporter transgenes to investigate the mechanisms of SLARP-mediated gene regulation.
In conclusion, GOMO is a robust selection procedure that allows the rapid isolation of isogenic drug-selectable marker-free parasites, in both P. berghei and P. yoelii, while drastically reducing animal usage. This strategy now expands the toolbox for gene targeting in rodent malaria parasites and will greatly facilitate the production of parasite mutants in the context of genetic screens.
Methods
Ethics statement
All animal work was conducted in strict accordance with the Directive 2010/63/EU of the European Parliament and Council ‘On the protection of animals used for scientific purposes’. The protocol was approved by the Charles Darwin Ethics Committee of the University Pierre et Marie Curie, Paris, France (permit number Ce5/2012/008).
Experimental animals, parasites and cell lines
Blood stage parasite infections were conducted in female Swiss mice (6–8 weeks old, from Janvier). Sporozoite infections were performed in female C57BL/6 mice (6 weeks old, from Janvier). We used the reference rodent malaria parasite strains P. berghei ANKA (clone 15cy1) and P. yoelii 17XNL (clone 1.1). Anopheles stephensi mosquitoes were infected by feeding on anesthetised infected mice using standard methods of mosquito infection27. P. berghei- and P. yoelii-infected mosquitoes were kept at 21°C and 24°C, respectively and fed daily on 10% sucrose. After 21 to 28 days (for P. berghei) or 14 to 18 days (for P. yoelii), the salivary glands of the mosquitoes were collected by hand-dissection and homogenized to collect sporozoites. HepG2 cells (ATCC HB-8065) were cultured in DMEM supplemented with 10% fetal calf serum and antibiotics, as described28.
Plasmid constructs
GOMO plasmids
Elements of the GOMO-GFP and GOMO-GFP-LUC plasmids were amplified by PCR, digested with appropriate restriction enzymes and sequentially ligated into a pBluescript backbone. We first assembled a bidirectional P. berghei eEF1α promoter fragment flanked by a mCherry coding sequence and a PbDHFR 3′ UTR fragment on one side and a hDHFR-yFCU coding sequence and a PbHSP70 3′ UTR fragment on the other. We then added either a GFP coding sequence under control of PbHSP70 promoter (GOMO-GFP vector), or a GFP-LUC coding sequence under control of P. berghei eEF1α promoter (GOMO-GFP-LUC). In each plasmid, the GFP or GFP-LUC coding sequence is followed by a sequence element corresponding to the PbDHFR 3′ UTR fragment, in the same orientation as the identical fragment placed downstream of mCherry (Fig. S1), allowing homologous recombination and concomitant excision of both the hDHFR-yFCU and mCherry cassettes. The GOMO plasmids were verified by Sanger sequencing (GATC Biotech).
Gene targeting vectors
For each target gene, a 5′ and a 3′ homology fragments were amplified by PCR and cloned into SacII/NotI and XhoI/KpnI sites, respectively, of GOMO-GFP and GOMO-GFP-LUC plasmids. The resulting vectors were then linearized with SacII and KpnI before transfection. All the primers used in the study are listed in Table S1.
Parasite transfection and selection procedure
WT P. berghei ANKA or P. yoelii 17XNL purified schizonts were transfected with 5–10 μg of linearized plasmid by electroporation using the AMAXA Nucleofector™ device (program U33), as described29 and immediately injected intravenously in the tail of a mouse. The day after transfection, pyrimethamine (70 mg/l or 7 mg/l for P. berghei or P. yoelii, respectively) was administered in the mouse drinking water, for positive selection of transgenic parasites. GFP+ mCherry+ pyrimethamine-resistant parasites (200 to 1,000) were then isolated by FACS and transferred into a naive mouse. Once the parasitemia reached 1%, 5-fluorocytosine (Meda Pharma) was added in the mouse drinking water at 1 mg/ml, as described9, for negative selection. Once the parasitemia reached 0.1–1%, GFP+ mCherry− parasites were sorted by flow cytometry and 20 to 50 parasites were injected intravenously into a recipient naive mouse to isolate pure isogenic marker-free recombinant parasite populations. FACS sorting of fluorescent parasites was performed on a FACSAria II (Becton-Dickinson), essentially as described6. Briefly, one drop of tail blood was collected and resuspended in 1 ml of Alsever's solution (Sigma–Aldrich), then passed through a 40 μm cell strainer (Falcon) to remove cell aggregates. Sorting of parasitized erythrocytes was performed using a purity sort-mask. Excitation of GFP and mCherry was done at wavelengths of 488 and 561 nm, respectively, while fluorescence emission was detected using band pass filters of 530/30 and 610/20 nm, respectively. Area and height of forward scatter signal were used to exclude doublets. Forward and sideward scatter gating was used to exclude small particles and overly large cells (Fig. S2). Gated GFP+ mCherry+ cells (first step) or GFP+ mCherry− cells (second step) were sorted and recovered in 200 μl of RPMI supplemented with 20% FCS and shortly injected into the tail vein of a naive mouse. Once the parasitemia reached more than 1%, the mouse blood was collected and either frozen or used for parasite genomic DNA isolation.
Genotyping
Infected blood was passed through a CF11 column (Whatman) to deplete leukocytes and erythrocytes were lysed with 0.2% saponin (Sigma) to remove haemoglobin. Parasite genomic DNA was extracted using the Purelink Genomic DNA Kit (Invitrogen) and analysed by PCR using primer combinations specific for WT and recombined loci (Table S1). Standard Southern blot analysis was performed using the PCR DIG probe synthesis kit and the DIG luminescence detection kit (Roche). Digoxigenin-labelled probes were synthesized with the same primers as used to produce PbP230p and PbSLARP gene replacement 3′ homology fragments (Table S1).
Sporozoite infection assays
For time lapse in vitro imaging, HepG2 cells (200,000 in a collagen-coated 24-well plate) were infected with 20,000 Δp230p-GFP or Δslarp-GFP P. berghei purified sporozoites and incubated for 90 min at 37°C. Infected cultures were then trypsinized and washed to remove all extracellular parasites and re-plated in a 96-well plate. Infected cells were imaged at 37°C and 5% CO2 from 7 to 24 hours post-infection, with a Zeiss Axio Observer.Z1 fluorescence microscope equipped with a LD Plan-Neofluar 40×/0.6 Corr Ph2 M27 objective. GFP and transmitted light images were captured every 20 min and processed with ImageJ for adjustment of contrast.
For in vivo imaging, C57BL/6 mice (n = 5 per group) were injected intravenously with 50,000 Δp230p-GFP-LUC or Δslarp-GFP-LUC P. berghei purified sporozoites. An additional group of uninfected mice (n = 3) was used as control. At 24, 48 and 96 hours post-infection, mice were injected intraperitoneally with 100 μl of a 30 mg/ml D-luciferin solution (Synchem, Germany) and anesthetized with 2% isoflurane. Bioluminescence images of whole mouse bodies were acquired with the IVIS Spectrum Imaging System (Caliper Life Sciences), 10 min after injection, with an exposure time of 3 to 60 seconds. Analysis was performed using Living Image, version 4.0 (Caliper Life Sciences) by measurement of photon flux (measured in photons/s/cm2/steradian).
Statistical analyses
Statistical significance was assessed by non-parametric analysis using the Mann-Whitney U and log rank (Mantel-Cox) tests. All statistical tests were computed with GraphPad Prism 5 (GraphPad Software).
References
Khan, S. M., Kroeze, H., Franke-Fayard, B. & Janse, C. J. Standardization in generating and reporting genetically modified rodent malaria parasites: the RMgmDB database. Methods Mol Biol 923, 139–50 (2013).
Heussler, V. & Doerig, C. In vivo imaging enters parasitology. Trends Parasitol 22, 192–5; discussion 195–6 (2006).
Ménard, R. & Janse, C. Gene targeting in malaria parasites. Methods 13, 148–57 (1997).
Philip, N., Orr, R. & Waters, A. P. Transfection of rodent malaria parasites. Methods Mol Biol 923, 99–125 (2013).
Janse, C. J. et al. High efficiency transfection of Plasmodium berghei facilitates novel selection procedures. Mol Biochem Parasitol 145, 60–70 (2006).
Kenthirapalan, S., Waters, A. P., Matuschewski, K. & Kooij, T. W. Flow cytometry-assisted rapid isolation of recombinant Plasmodium berghei parasites exemplified by functional analysis of aquaglyceroporin. Int J Parasitol 42, 1185–92 (2012).
de Koning-Ward, T. F. et al. The selectable marker human dihydrofolate reductase enables sequential genetic manipulation of the Plasmodium berghei genome. Mol Biochem Parasitol 106, 199–212 (2000).
Braks, J. A., Franke-Fayard, B., Kroeze, H., Janse, C. J. & Waters, A. P. Development and application of a positive-negative selectable marker system for use in reverse genetics in Plasmodium. Nucleic Acids Res 34, e39 (2006).
Orr, R. Y., Philip, N. & Waters, A. P. Improved negative selection protocol for Plasmodium berghei in the rodent malarial model. Malar J 11, 103 (2012).
Lin, J. W. et al. A novel ‘gene insertion/marker out’ (GIMO) method for transgene expression and gene complementation in rodent malaria parasites. PLoS One 6, e29289 (2011).
Kooij, T. W., Rauch, M. M. & Matuschewski, K. Expansion of experimental genetics approaches for Plasmodium berghei with versatile transfection vectors. Mol Biochem Parasitol 185, 19–26 (2012).
Amino, R. et al. Host cell traversal is important for progression of the malaria parasite through the dermis to the liver. Cell Host Microbe 3, 88–96 (2008).
Mwakingwe, A. et al. Noninvasive real-time monitoring of liver-stage development of bioluminescent Plasmodium parasites. J Infect Dis 200, 1470–8 (2009).
Ploemen, I. H. et al. Visualisation and quantitative analysis of the rodent malaria liver stage by real time imaging. PLoS One 4, e7881 (2009).
van Dijk, M. R. et al. Three members of the 6-cys protein family of Plasmodium play a role in gamete fertility. PLoS Pathog 6, e1000853 (2010).
Hliscs, M., Nahar, C., Frischknecht, F. & Matuschewski, K. Expression profiling of Plasmodium berghei HSP70 genes for generation of bright red fluorescent parasites. PLoS One 8, e72771 (2013).
Aly, A. S. et al. Targeted deletion of SAP1 abolishes the expression of infectivity factors necessary for successful malaria parasite liver infection. Mol Microbiol 69, 152–63 (2008).
Silvie, O., Goetz, K. & Matuschewski, K. A sporozoite asparagine-rich protein controls initiation of Plasmodium liver stage development. PLoS Pathog 4, e1000086 (2008).
van Dijk, M. R. et al. Genetically attenuated, P36p-deficient malarial sporozoites induce protective immunity and apoptosis of infected liver cells. Proc Natl Acad Sci U S A 102, 12194–9 (2005).
Hafalla, J. C., Silvie, O. & Matuschewski, K. Cell biology and immunology of malaria. Immunol Rev 240, 297–316 (2011).
Silvie, O. et al. Hepatocyte CD81 is required for Plasmodium falciparum and Plasmodium yoelii sporozoite infectivity. Nat Med 9, 93–6 (2003).
Maier, A. G., Braks, J. A., Waters, A. P. & Cowman, A. F. Negative selection using yeast cytosine deaminase/uracil phosphoribosyl transferase in Plasmodium falciparum for targeted gene deletion by double crossover recombination. Mol Biochem Parasitol 150, 118–21 (2006).
Pfander, C. et al. A scalable pipeline for highly effective genetic modification of a malaria parasite. Nat Methods 8, 1078–82 (2011).
Mueller, A. K. et al. Plasmodium liver stage developmental arrest by depletion of a protein at the parasite-host interface. Proc Natl Acad Sci U S A 102, 3022–7 (2005).
Mueller, A. K., Labaied, M., Kappe, S. H. & Matuschewski, K. Genetically modified Plasmodium parasites as a protective experimental malaria vaccine. Nature 433, 164–7 (2005).
Silvie, O. et al. Effects of irradiation on Plasmodium falciparum sporozoite hepatic development: implications for the design of pre-erythrocytic malaria vaccines. Parasite Immunol 24, 221–3 (2002).
Ramakrishnan, C. et al. Laboratory maintenance of rodent malaria parasites. Methods Mol Biol 923, 51–72 (2013).
Silvie, O., Franetich, J. F., Boucheix, C., Rubinstein, E. & Mazier, D. Alternative invasion pathways for Plasmodium berghei sporozoites. Int J Parasitol 37, 173–82 (2007).
Janse, C. J., Ramesar, J. & Waters, A. P. High-efficiency transfection and drug selection of genetically transformed blood stages of the rodent malaria parasite Plasmodium berghei. Nat Protoc 1, 346–56 (2006).
Acknowledgements
We would like to thank Thierry Houpert and Maurel Tefit for mosquito rearing and the personnel of the Centre d'Expérimentation Fonctionnelle (CEF) of Pitié-Salpêtrière for taking care of the mice. This work was funded by the European Union (FP7 Marie Curie grant PCIG10-GA-2011-304081), the Agence Nationale de la Recherche (ANR-10-PDOC-008-01), the National Centre for the Replacement, Refinement and Reduction of Animals in Research (NC/L000601/1) and the Laboratoire d'Excellence (LabEx) ParaFrap (ANR-11-LABX-0024). G.M. was supported by a “DIM Malinf” doctoral fellowship awarded by the Conseil Régional d'Ile-de-France. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
G.M. performed experiments, analysed data and wrote the manuscript. S.B. performed experiments and analysed data. V.R.C., C.G., S.T. and M.L.I. performed experiments. J.F.F. supervised mosquito production. B.H. performed flow cytometry. D.M. analysed data. O.S. designed experiments, analysed data and wrote the manuscript. All authors reviewed the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary Information
Supplementary Information
Supplementary movie 1
Supplementary Information
Supplementary movie 2
Supplementary Information
Supplementary movie 3
Supplementary Information
Supplementary movie 4
Supplementary Information
Supplementary movie 5
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Unported License. The images in this article are included in the article's Creative Commons license, unless indicated otherwise in the image credit; if the image is not included under the Creative Commons license, users will need to obtain permission from the license holder in order to reproduce the image. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Manzoni, G., Briquet, S., Risco-Castillo, V. et al. A rapid and robust selection procedure for generating drug-selectable marker-free recombinant malaria parasites. Sci Rep 4, 4760 (2014). https://doi.org/10.1038/srep04760
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep04760
This article is cited by
-
Gene copy number and function of the APL1 immune factor changed during Anopheles evolution
Parasites & Vectors (2020)
-
Prediction of malaria transmission drivers in Anopheles mosquitoes using artificial intelligence coupled to MALDI-TOF mass spectrometry
Scientific Reports (2020)
-
Improvement of an in vitro drug selection method for generating transgenic Plasmodium berghei parasites
Malaria Journal (2019)
-
Differential activity of methylene blue against erythrocytic and hepatic stages of Plasmodium
Malaria Journal (2018)
-
Cytotoxic anti-circumsporozoite antibodies target malaria sporozoites in the host skin
Nature Microbiology (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.