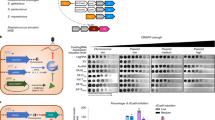
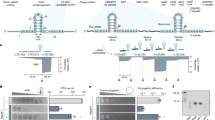
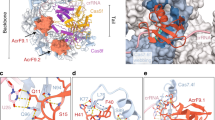
Abstract
CRISPR-Cas systems provide sequence-specific adaptive immunity against foreign nucleic acids1,2. They are present in approximately half of all sequenced prokaryotes3 and are expected to constitute a major barrier to horizontal gene transfer. We previously described nine distinct families of proteins encoded in Pseudomonas phage genomes that inhibit CRISPR-Cas function4,5. We have developed a bioinformatic approach that enabled us to discover additional anti-CRISPR proteins encoded in phages and other mobile genetic elements of diverse bacterial species. We show that five previously undiscovered families of anti-CRISPRs inhibit the type I-F CRISPR-Cas systems of both Pseudomonas aeruginosa and Pectobacterium atrosepticum, and a dual specificity anti-CRISPR inactivates both type I-F and I-E CRISPR-Cas systems. Mirroring the distribution of the CRISPR-Cas systems they inactivate, these anti-CRISPRs were found in species distributed broadly across the phylum Proteobacteria. Importantly, anti-CRISPRs originating from species with divergent type I-F CRISPR-Cas systems were able to inhibit the two systems we tested, highlighting their broad specificity. These results suggest that all type I-F CRISPR-Cas systems are vulnerable to inhibition by anti-CRISPRs. Given the widespread occurrence and promiscuous activity of the anti-CRISPRs described here, we propose that anti-CRISPRs play an influential role in facilitating the movement of DNA between prokaryotes by breaching the barrier imposed by CRISPR-Cas systems.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Brouns, S. J. et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321, 960–964 (2008).
Barrangou, R. et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712 (2007).
Makarova, K. S. et al. An updated evolutionary classification of CRISPR-Cas systems. Nature Rev. Microbiol. 13, 722–736 (2015).
Bondy-Denomy, J., Pawluk, A., Maxwell, K. L. & Davidson, A. R. Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature 493, 429–432 (2013).
Pawluk, A., Bondy-Denomy, J., Cheung, V. H., Maxwell, K. L. & Davidson, A. R. A new group of phage anti-CRISPR genes inhibits the type I-E CRISPR-Cas system of Pseudomonas aeruginosa. mBio 5, e00896 (2014).
Doolittle, W. F. Phylogenetic classification and the universal tree. Science 284, 2124–2129 (1999).
Gogarten, J. P., Doolittle, W. F. & Lawrence, J. G. Prokaryotic evolution in light of gene transfer. Mol. Biol. Evol. 19, 2226–2238 (2002).
Marraffini, L. A. & Sontheimer, E. J. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322, 1843–1845 (2008).
Garneau, J. E. et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468, 67–71 (2010).
Jore, M. M. et al. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nature Struct. Mol. Biol. 18, 529–536 (2011).
Wiedenheft, B. et al. Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature 477, 486–489 (2011).
Westra, E. R. et al. CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Mol. Cell 46, 595–605 (2012).
Hale, C. R. et al. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell 139, 945–956 (2009).
Gophna, U. et al. No evidence of inhibition of horizontal gene transfer by CRISPR-Cas on evolutionary timescales. ISME J. 9, 2021–2027 (2015).
Koonin, E. V., Makarova, K. S. & Aravind, L. Horizontal gene transfer in prokaryotes: quantification and classification. Annu. Rev. Microbiol. 55, 709–742 (2001).
Smillie, C. S. et al. Ecology drives a global network of gene exchange connecting the human microbiome. Nature 480, 241–244 (2011).
Bondy-Denomy, J. et al. Multiple mechanisms for CRISPR-Cas inhibition by anti-CRISPR proteins. Nature 526, 136–139 (2015).
Wiedenheft, B. In defense of phage: viral suppressors of CRISPR-mediated adaptive immunity in bacteria. RNA Biol. 10, 886–890 (2013).
Richter, C., Gristwood, T., Clulow, J. S. & Fineran, P. C. PLoS ONE 7, e49549 (2012).
Przybilski, R. et al. Csy4 is responsible for CRISPR RNA processing in Pectobacterium atrosepticum. RNA Biol. 8, 517–528 (2011).
Vercoe, R. B. et al. Cytotoxic chromosomal targeting by CRISPR/Cas systems can reshape bacterial genomes and expel or remodel pathogenicity islands. PLoS Genet. 9, e1003454 (2013).
Van Belkum, A. et al. Phylogenetic distribution of CRISPR-Cas systems in antibiotic-resistant Pseudomonas aeruginosa. mBio 6, 01796-15 (2015).
Altschul, S. F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997).
Katoh, K., Misawa, K., Kuma, K. & Miyata, T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066 (2002).
Waterhouse, A. M., Procter, J. B., Martin, D. M., Clamp, M. & Barton, G. J. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 (2009).
Qiu, D., Damron, F. H., Mima, T., Schweizer, H. P. & Yu, H. D. PBAD-based shuttle vectors for functional analysis of toxic and highly regulated genes in Pseudomonas and Burkholderia spp. and other bacteria. Appl. Environ. Microbiol. 74, 7422–7426 (2008).
Cady, K. C., Bondy-Denomy, J., Heussler, G. E., Davidson, A. R. & O'Toole, G. A. The CRISPR/Cas adaptive immune system of Pseudomonas aeruginosa mediates resistance to naturally occurring and engineered phages. J. Bacteriol. 194, 5728–5738 (2012).
Strotksaya, A., Semenova, E., Savitskaya, E. & Severinov, K. Rapid multiplex creation of Escherichia coli strains capable of interfering with phage infection through CRISPR. Methods Mol. Biol. 1311, 147–159 (2015).
Richter, C. et al. Priming in the Type I-F CRISPR-Cas system triggers strand-independent spacer acquisition, bi-directionally from the primed protospacer. Nucleic Acids Res. 42, 8516–8526 (2014).
Blower, T. R., Evans, T. J., Przybilski, R., Fineran, P. C. & Salmond, G. P. Viral evasion of a bacterial suicide system by RNA-based molecular mimicry enables infectious altruism. PLoS Genet. 8, e1003023 (2012).
Acknowledgements
This work was supported by funding from the Canadian Institutes of Health Research to A.R.D. (MOP-130482) and K.L.M. (MOP-136845). A.P. was supported by an Ontario Graduate Scholarship and a CIHR Canada Graduate Scholarship Doctoral Award. R.H.J.S. was funded by a University of Otago Division of Health Sciences Career Development Post-doctoral Fellowship. P.C.F was supported by a Rutherford Discovery Fellowship from the Royal Society of New Zealand. B.N.J.W. was supported by a University of Otago Doctoral Scholarship.
Author information
Authors and Affiliations
Contributions
A.P. designed experiments, performed bioinformatic analysis, performed experiments, analysed data and wrote the manuscript. R.H.J.S. designed and performed Pectobacterium experiments and wrote parts of the manuscript. C.T. performed Pectobacterium experiments. B.N.J.W. performed Pectobacterium experiments and wrote parts of the manuscript. S.S. assembled and uploaded to NCBI the genome sequences of P. aeruginosa strains SMC4386 and SMC4386 ΔCRISPR-Cas. P.C.F. designed Pectobacterium experiments and wrote parts of the manuscript. K.L.M. supervised experiments and wrote the manuscript. A.R.D. supervised experiments and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Figure 1, Supplementary Tables 1-6, Supplementary References. (PDF 514 kb)
Rights and permissions
About this article
Cite this article
Pawluk, A., Staals, R., Taylor, C. et al. Inactivation of CRISPR-Cas systems by anti-CRISPR proteins in diverse bacterial species. Nat Microbiol 1, 16085 (2016). https://doi.org/10.1038/nmicrobiol.2016.85
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/nmicrobiol.2016.85
This article is cited by
-
Viruses wrap up bacterial defence systems
Nature (2024)
-
Associate toxin-antitoxin with CRISPR-Cas to kill multidrug-resistant pathogens
Nature Communications (2023)
-
Bacteriophage therapy: an emerging paradigm in fish disease management
Aquaculture International (2023)
-
Phylogenetic Analysis of Anti-CRISPR and Member Addition in the Families
Molecular Biotechnology (2023)
-
Anti-CRISPR proteins: a weapon of phage-bacterial arm race for genome editing
The Nucleus (2023)