Abstract
Recent advances in the development of high-throughput genotyping platforms allow for the unbiased identification of genes and genomic sequences related to heritable traits. In this study, we analyzed human short-term memory, which refers to the ability to remember information over a brief period of time and which has been found disturbed in many neuropsychiatric conditions, including schizophrenia and depression. We performed a genome-wide survey at 909 622 polymorphic loci and report six genetic variations significantly associated with human short-term memory performance after genome-wide correction for multiple comparisons. A polymorphism within SCN1A (encoding the α subunit of the type I voltage-gated sodium channel) was replicated in three independent populations of 1699 individuals. Functional magnetic resonance imaging during an n-back working memory task detected SCN1A allele-dependent activation differences in brain regions typically involved in working memory processes. These results suggest an important role for SCN1A in human short-term memory.
Similar content being viewed by others
Introduction
The search for molecules related to cognitive traits such as human memory functions is key for the understanding of the underlying biology and for the identification of molecular pathways possibly amenable to pharmacologic intervention in pathological conditions. Unlike candidate-gene association studies, which depend on pre-existing information,1, 2, 3, 4 recent advances in the development of high-density genotyping platforms now allow for high-resolution genome-wide association studies (GWAS) of polygenic phenotypes resulting in the identification of novel genes and genomic sequences related to cognitive traits.5
In this study, we used an individual genome-wide screen with more than 900 000 single-nucleotide polymorphisms (SNPs) in 333 Swiss healthy young individuals to identify genetic loci related to human short-term memory processes as assessed by a verbal immediate recall task. SNPs reaching statistically significant levels after genome-wide correction for multiple comparisons were analyzed in a second cohort of 254 Swiss healthy young individuals who performed an identical immediate recall task (phase 2). Significant phase 2 SNPs were further analyzed in 922 elderly, non-demented individuals from Germany (phase 3) and in 523 healthy young individuals from Serbia (phase 4) who had also performed verbal immediate recall tasks. To further validate and characterize the findings of the behavioral genetics studies, we used functional magnetic resonance imaging (fMRI), which can detect genotype-dependent differences in brain activations during cognitive tasks.2, 3, 6
Materials and methods
Memory testing
Swiss sample (phases 1 and 2)
We recruited 333 (phase 1) and 254 (phase 2) healthy, young Swiss university students or age-matched employees/trainees. Of the 587 participants of phases 1 and 2, 426 were women and 161 were men. Age was 22.1±3.2 years (mean±s.d.). After complete description of the study to the participants, written informed consent was obtained. The ethics committee of the Canton of Zurich, Switzerland approved the study protocol. Participants viewed six series of five semantically unrelated nouns presented at a rate of one word per second with the instruction to learn the words for immediate free recall after each series. The number of correctly recalled words (hits) was the relevant output. All participants were tested with a standardized mental health questionnaire for current and life-time major psychiatric diseases, smoking and the use of any medication. In addition, participants provided information on possible familiar aggregation of major psychiatric diseases, female participants gave also information on the use of oral contraceptives and the actual menstrual cycle. Blood was drawn by using 2 × 9 ml ethylene-diaminetetraacetic acid tubes (Sarstedt, Germany), DNA isolation was carried out according to standard protocols (Qiagen, Hilden, Germany).
German sample (phase 3)
Participants were drawn from a large population-based cohort of elderly individuals participating in the AgeCoDe study. This study follows 3327 non-demented, non-institutionalized participants aged 75–90 years recruited through general practitioners in six German cities. The design of the AgeCoDe study is given in more detail elsewhere.7, 8 The ethics committees of the participating centers approved the study. Written informed consent was obtained from all participants. From the full sample with available DNA we excluded all participants where health conditions, medication or other factors could significantly affect cognitive performance. The exclusion criteria were: visual or auditory impairments possibly limiting the cognitive evaluation, neurological disorders (for example, Parkinson Disease, seizures, transitory ischemic attacks, stroke, closed head injury with more than 30 min unconsciousness), diabetes, current depression (Geriatric Depression scale ⩾6) or intake of tricyclic antidepressants, poor motivation during testing, first language other than German, harmful use of alcohol or alcohol addiction, or medication with benzodiazepines. We included 922 participants in this study.
Neuropsychological testing
All participants were assessed at their home by trained research assistants with a battery of tests. These include a structured interview for the assessment of dementia, the word list learning task (episodic memory) and the category fluency task of the consortium to establish a registry for Alzheimer's disease battery.9 The total number of words recalled after the first learning trial (maximum=10) was used as a measure of verbal short-term memory.
Serbian sample (phase 4)
Study participants were 523 healthy young individuals (351 female and 172 male students of the School of Medicine of the University of Belgrade, age 20.3±0.9 years) who agreed to participate in a study of the genetic basis of human cognition. Free recall of a short story narrative (immediate and 30 min delayed recall of items from Story A of the Logical Memory test; Wechsler Memory Scale-revised)10 was used to assess verbal memory performance. Participants completed also a questionnaire related to general health, living circumstances, parental education and subjective learning capacity.
Statistics and single-nucleotide polymorphism selection
The GWAS (phase 1) and the phase 2 replication studies were run under the assumption of an additive model (trend test). False discovery rate (FDR) was used to correct for genome-wide multiple testing in phase 1. The FDR-corrected significance level (P1_FDR) was set to 0.05. After initial quality control (QC), we used following criteria to identify significant phase 1 SNPs to follow in phase 2: (a) P1_FDR⩽0.05; (b) minor allele frequency ⩾0.05; (c) Hardy–Weinberg equilibrium P⩾0.05; (d) intragenic SNP localization (that is, within genes/open-reading frames or within a haplotype block extending to an adjacent gene/open-reading frame). Of the 650 SNPs fulfilling criterion (a) only six SNPs fulfilled the additional criteria (b) through (d). Genomic control analysis using all SNP data was used to calculate the inflation factor λ, which was 1.0 in the homogenous Swiss sample from phase 1. Thus, the significances reported herein are not affected by stratification.
The haplotype trend regression method was used for haplotype analysis. Maximum expectation maximization iterations and expectation maximization convergence tolerance were set to 50 and 0.0001, respectively. Minimum haplotype frequency was set to 5%. The computed significance applied to the full model, that is, haplotypes+non-haplotype covariates.
In the German sample of elderly individuals, analysis of variance was used to calculate the significance of association between SNPs and memory performance. Each analysis was corrected for age, gender, the presence of mild cognitive impairment, and education. In the Serbian sample of young students, analysis of variance corrected for gender and living circumstances effects was used to calculate the significance of association between SNP rs10930201 and memory performance.
Helix Tree (version 6.4.1, Golden Helix Inc., Bozeman, MT, USA) and SPSS 11 (SPSS Inc., Chicago, IL, USA) were used for statistical analyses.
Array-based single-nucleotide polymorphism genotyping
Samples were processed as described in the Genome-Wide Human SNP Nsp/Sty 6.0 User Guide (Affymetrix Inc., Santa Clara, CA, USA). Briefly, genomic DNA concentration was determined by using a Nano-Drop ND-1000 and adjusted to 50 ng μl–1 in water. In all, 250 ng of DNA was digested in parallel with 10 units of Sty I and Nsp I restriction enzymes (New England Biolabs, Beverly, MA, USA) for 2 h at 37 °C. Enzyme-specific adapter oligonucleotides were then ligated onto the digested ends with T4 DNA Ligase for 3 h at 16 °C. After adjustment to 100 μl with water, 10 μl of the diluted ligation reactions were subjected to PCR. Three PCR reactions of 100 μl were performed for Sty-digested products and four PCR reactions for Nsp. PCR was performed using Titanium Taq DNA Polymerase (Clontech, Mountain View, CA, USA) in the presence of 4.5 μM PCR primer 002 (Affymetrix), 350 μM each dNTP (Clontech), 1 M G-C Melt (Clontech) and 1 × Titanium Taq PCR Buffer (Clontech). Cycling parameters were as follows, initial denaturation at 94 °C for 3 min, amplification at 94 °C for 30 s, 60 °C for 45 s and extension at 68 °C for 15 s repeated a total of 30 times, final extension at 68 °C for 7 min. Reactions were then verified to migrate at an average size between 200 and 1100 bp using 2% TBE gel electrophoresis. PCR products were combined and purified with the Filter Bottom Plate (Seahorse Bioscience, North Billerica, MA, USA) using Agencourt Magnetic Beads (Beckman Coulter, Fullerton, CA, USA). Purified PCR products were quantified on a Zenith 200rt microplate reader (Anthos-Labtec, Cambridge, UK). In all, 4–5 μg μl–1 were obtained on average for each sample. From this stage on, the SNP Nsp/Sty 5.0/6.0 Assay Kit (Affymetrix) was used. Around 250 μg of purified PCR products were fragmented using 0.5 units of DNAse I at 37 °C for 35 min. Fragmentation of the products to an average size <180 bp was verified using 4% TBE gel electrophoresis. After fragmentation, the DNA was end labeled with 105 units of terminal deoxynucleotidyl transferase at 37 °C for 4 h. The labeled DNA was then hybridized onto Genome-Wide Human SNP 6.0 Array at 50 °C for 18 h at 60 revolutions per minute. The hybridized array was washed, stained, and scanned according to the manufacturer's (Affymetrix) instructions using Affymetrix GeneChip Command Console (AGCC, version 0.0.0.676). Generation of SNP calls and array quality control were performed using the Affymetrix Genotyping Console Software 3.0 (Affymetrix Inc.). According to the manufacturer's recommendation, Contrast QC was chosen as QC metric, using the default value of ⩾0.4. Individual QC Call Rate Threshold was set to default of ⩾0.86. Mean Call Rate for all samples averaged 98.7%. All samples passing QC criteria were subsequently genotyped using the Birdseed (v1) algorithm. A total of 909 622 SNPs were genotyped and imported into Helix Tree (version 6.4.1). For association testing 67 929 markers with fewer than two alleles and 30 markers with insufficient cardinality were skipped leaving a total of 841 663 markers to be analyzed.
Singleplex single-nucleotide polymorphism genotyping
Singleplex genotyping was carried out with Pyrosequencing on a PyroMarkID System (Biotage, Uppsala, Sweden). The chip-based SNP_A-8386800 (rs10930201) was also genotyped by Pyrosequencing in the samples from phase 1 and phase 2. In the phase 1 sample, the level of convergence between chip- and Pyrosequencing-based genotype calls was 100%. The non-synonymous SNP rs2298771 was also genotyped by Pyrosequencing. Please contact the authors for the detailed list of singleplex primers.
Magnetic resonance imaging
Participants
In total, 9 men and 15 women; median age 22 years, range 18–25 years.
Working memory functional magnetic resonance imaging experiment
There was a single fMRI time-series with a two-back task assessing working memory and a baseline task (‘x-target’ task) measuring concentration. The two-back task required participants to respond to a letter repeat with one intervening letter (for example, S−f−s−g…). The ‘x-target’ task required participants to respond to the occurrence of the letter ‘x’ in a sequence of letters (for example, N−l−X−g…). We presented 50 upper- or lowercase letters typed in black on white background. The use of upper- and lowercase letters prevented a visual-matching strategy. There were five blocks of 26 s per condition. Each block comprised 13 upper- or lowercase letters, which were presented for 2 s each.
Data acquisition
Magnetic resonance measurements were performed using a 3 T Philips Intera whole-body magnetic resonance scanner equipped with an eight-channel Philips (Best, The Netherlands) SENSE head coil. Functional data were obtained from 32 transverse slices parallel to the anterior commissure-posterior commissure plane covering the whole brain with a measured spatial resolution of 2.8 × 2.8 × 4 mm3 (acquisition matrix 80 × 80) and a reconstructed resolution of 1.7 × 1.7 × 4 mm3. Data were acquired with a SENSE-sshEPI sequence with an acceleration factor of R=2.0.11 Other scan parameters were echo time (TE)=35 ms, repetition time (TR)=3000 ms, θ=82°. A 3D T1-weighted anatomical scan was obtained for the voxel-based morphometry analysis with a measured spatial resolution of 1 × 1 × 1.5 mm3 (acquisition matrix 224 × 224) and a reconstructed resolution of 0.9 × 0.9 × 0.8 mm3, TE=2.3 ms, TR=20 ms, θ=20°, no interslice gaps.
Analysis of functional magnetic resonance imaging data
Image pre- and postprocessing and the statistical analyses were performed using SPM2. Standard preprocessing procedures were applied, realignment, normalization and spatial smoothing (8 mm). On the single subject level, data were analyzed according to the fixed effects model (SPM2; http://www.fil.ion.ucl.ac.uk/spm/). Contrasts were computed for each subject comparing the two-back task to the x-target task. At the second level, within-subject contrasts were entered into random effects analyses (Two sample t-tests, SPM2)12 and correlated with behavioral performance (simple regression, SPM2). The six head movement parameters were included in the model as confounding factors. Data were high-pass filtered with a filter-value tailored to each fMRI time-series according to 2 × stimulus onset asynchrony × TR. Threshold was set at a P<0.001 level, uncorrected for multiple comparisons, with an extent threshold of 20 voxels. SPM2 coordinates refer to standard brains from the Montreal Neurological Institute.
Analysis of anatomical magnetic resonance imaging data
Image preprocessing was carried out with Statistical Parametric Mapping (SPM2) software (http://www.fil.ion.ucl.ac.uk/spm) using optimized voxel-based morphometry13, 14 implemented in scripts written by C Gaser (http://dbm.neuro.uni-jena.de/vbm/) that computes voxel-wise comparisons of gray matter, white matter and liquor probabilities between groups. We normalized and segmented the native magnetic resonance images using the MNI 152 a priori maps. In additional, we modulated the gray matter images with the Jacobian determinant that account for volumetric changes introduced by the spatial normalization. These gray matter images were then smoothed with a 12 mm Gaussian kernel and subjected into a voxel-wise statistical analysis.
Data analysis of cortical gene expression data
Data are based on the survey of genetic human cortical gene expression published recently by Myers et al.15 Gene expression studies of 178 samples from the cerebral cortex of neurpathologically normal brains were carried out with the Illunima HumanRefseq-8 Expression BeadChip (Illumina Inc., San Diego, CA, USA). For genome-wide genotyping, the Affymetrix GeneChip Human Mapping 500 K Array Set was used. The complete data files were downloaded from http://labs.med.miami.edu/myers/. SCN1A transcript probe was GI_29893558 (NM_006920.2) and expression levels of GI_29893558 were used as dependent variable. The genetic association analysis was run under the assumption of an additive model (trend test), which was also used tor the GWAS on memory performance.
Results
Genome-wide survey and replication studies
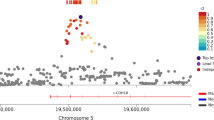
Participants (phase 1) viewed six series of five semantically unrelated nouns presented at a rate of one word per second with the instruction to learn the words for immediate free recall after each series, resulting in a recall of totally 24.0±0.2 (mean±s.e.m.) words. Individual genotyping with the Affymetrix Genome-Wide Human SNP Array 6.0 showed that six common SNPs were significantly associated with immediate recall performance also after genome-wide correction (FDR) for multiple comparisons (Figure 1) (see Materials and Methods section). The significant SNPs from phase 1 (n=6) were genotyped in a second cohort of 254 healthy individuals who performed the identical immediate recall task. In this group, mean immediate recall was 23.5±0.2 words. SNP rs10930201, located within the gene encoding voltage-gated sodium channel type 1a (SCN1A), was significantly associated with immediate recall also in the phase 2 sample (Figure 1 and Supplementary Figure 1). In both samples, carriers of the minor allele presented with poorer short-term memory performance in an allele dose-dependent manner (Table 1). This result was also confirmed in the phase 3 sample (P=0.03), which consisted of 922 non-demented elderly individuals (mean age: 80 years, range: 75–90 years) and in the phase 4 sample (P=0.0002, Table 1), which consisted of 523 healthy young individuals (mean age: 20 years, range: 19–27 years) (see Materials and Methods section). In the samples from phases 1, 2 and 4, we observed no significant association of SNP rs10930201 with delayed recall of the presented material (phases 1 and 2: 5 min and 24 h delayed recall; phase 4: 30 min delayed recall; all Ps>0.1). In the phase 3 sample of elderly individuals, this SNP was associated (in the same allele-dependent manner) with delayed recall performance (10 min delayed recall, P=0.006). However, after correction for immediate recall performance, the association between rs10930201 and delayed recall failed to reach significance (P>0.05), indicating a carry-over genotype effect from the immediate to the delayed recall test. These findings suggest that the SCN1A genotype is mainly related to short-term memory processes as measured by immediate recall tests.
Genome-wide association and replication. Upper panel: genome-wide results (−log10P), corrected for genome-wide multiple comparisons (false discovery rate (FDR)), are shown in chromosomal order for individually genotyped single-nucleotide polymorphisms (SNPs) that were tested for association with immediate verbal recall performance in the phase 1 sample of 333 individuals. The red dotted line indicates the FDR-corrected significance level, which was exceeded by six SNPs. Lower panel: list of significant SNPs (FDR-corrected) with corresponding chromosomal positions and phase 1 significance values. Chromosomal positions were retrieved from the March 2006 UCSC genome browser assembly. MAF, minor allele frequency; r, genotype-phenotype correlation in phase 1 sample; P, uncorrected significance level in phase 1 sample. The additive model was used for all association tests. The color reproduction of this figure is available on the html full text version of the manuscript.
Analysis of all 39 SCN1A SNPs represented on the chip showed that nine intragenic SNPs were significantly associated with immediate recall performance in phase 1 (P<0.05, corrected for 39 SNPs, Supplementary Figure 1a). Haplotype analysis showed that a four SNP haplotype covering almost the entire gene and harboring the intronic SNP rs10930201 was significantly associated with memory performance (P=0.0000002) (Supplementary Figure 1a). In the phase 2 sample, this haplotype was significantly associated with memory performance as well (P=0.004, Supplementary Figure 1b). In the phase 1 sample, SNP rs6731900 located 1 kb 5′ to the TTC21B gene (encoding tetratricopeptide repeat domain 21B) and 103 kb 3′ to SCN1A SNP rs10930201 was also significantly (that is, FDR-corrected) associated with immediate recall performance (Figure 1). Although rs6731900 did not replicate in the phase 2 sample (Figure 1), we performed a post hoc analysis of all informative TTC21B SNPs represented on the 6.0 chip (n=10, minor allele frequency >0.05, Hardy–Weinberg equilibrium P>0.05) to rule out the possibility that TTC21B rather than SCN1A is associated with human memory performance. In the phase 1 sample, we did not observe any FDR-corrected significant TTC21B SNPs. In addition, no TTC21B SNPs reaching nominal significance were observed in the phase 2 sample (Supplementary Table 1).
Genetic variability within SCN1A is linked to human brain function
Immediate recall performance as assessed in this study consisted of recalling verbal information immediately after presentation and learning. This kind of short-term storage is an essential component of working memory.16, 17 To further analyze genotype-dependent differences in brain activation patterns, we used a verbal two-back task, which has been widely used in neuroimaging studies investigating the neural basis of working memory.18 Participants were required to monitor a series of visually presented letters on a trial-by-trial basis. They had to remember the last two letters in their temporal order, compare the currently presented letter with the one that occurred in the two-back task, and update their memory by dropping the oldest letter and incorporating the most recently presented letter. We applied the two-back letter task during fMRI in a sample of 24 healthy young individuals drawn from the phase 1 sample (12 homozygous carriers of the A allele of SNP rs10930201, 12 heterozygous AC carriers). The genotype groups were matched for sex (five men and seven women in the homozygous, four men and eight women in the heterozygous), age and immediate recall performance (all P-values>0.8). Performance was matched to avoid measuring genotype-unrelated performance effects on brain activations, and to instead capture genotype-dependent differences in brain activation patterns. As expected based on the matching for immediate recall performance on grounds of data collected in the large sample, there were no genotype-dependent differences in two-back task performance in the fMRI (P=0.5). The pooled data (over both genotype groups) showed robust activation in a bilateral fronto-parietal network (Supplementary Figure 2), including Brodmann area (BA) 6 and 44, as typically found in neuroimaging studies of working memory17 and as found in our previous study using the same task and magnetic resonance scanning protocol.6 During the two-back task (contrasted with ‘x-target’ baseline task) (see Materials and Methods section) heterozygous individuals showed significantly increased brain activations compared with homozygous A allele carriers in the right superior frontal gyrus/sulcus, BA 6 (local maximum at coordinate position (22, 18, 56), t=4.56, P<0.001; coordinates according to the Montreal Neurological Institute) (Figure 2a). This area has been consistently and bilaterally activated by n-back tasks, independently of stimulus type and identity versus location monitoring.18, 19 In a meta-analysis of working memory studies,20 this area responded when working memory needed to be continuously updated and when temporal order was crucial, which is true for n-back tasks. Although heterozygous individuals recruited more neural resources in BA 6, homozygous carriers of the A allele showed increased activity in the right inferior frontal sulcus, BA 44 (local maximum at coordinate position (48, 16, 22), t=4.29, P<0.001) (Figure 2b). BA 44 is typically activated during working memory for verbal stimuli,18, 20 also during n-back letter tasks,21, 22, 23, 24 and is thought to contribute to rehearsal or to the active maintenance of information within working memory.21, 22, 23, 24 Whereas most studies reported left lateralization, several studies also reported additional activations in the right BA 44, especially with increasing working memory load (for example, Braver et al.21; Cohen et al.22).
Significant SCN1A allele-dependent differences in brain activation during a two-back working memory task. (a) Heterozygous carriers of the minor allele C showed significantly increased brain activations compared with homozygous A allele carriers in the right superior frontal gyrus/sulcus (BA 6). (b) Homozygous A allele carriers showed significantly increased brain activations compared with heterozygous individuals in the right inferior frontal sulcus (BA 44). Activations were overlaid on sagittal, coronal and horizontal sections of a T1-weighted magnetic resonance image of SPM2 and showed in color-coded t-values. Threshold: P<0.001, uncorrected for multiple comparisons.
Studies investigating inter-individual differences in verbal working memory performance (based on both genetic and environmental factors) have indicated that high performers show lower brain activity in working memory-related brain regions than low performers.25, 26, 27 In the present fMRI experiment, however, the two genotype groups were performance matched in terms of verbal short-term memory capacity based on the data collected in the behavioral genetics cohort. This matching might have circumvented broader genotype-dependent activation differences within the working-memory network (for example, in the posterior parietal cortex) but rather revealed a differential activation pattern in BA 6 and BA 44.
Automated voxel-based morphometry (SPM2) (see Materials and Methods section) did not reveal significant genotype-dependent differences in gray matter nor white matter concentrations, indicating that functional imaging results were not biased by morphological differences.
Discussion
By using an unbiased, genome-wide behavioral genetics approach combined with functional brain imaging studies, we present evidence suggesting that the α subunit of the voltage-gated sodium channel, type I (protein: Nav1. 1; gene: SCN1A) has a significant role in human short-term memory. Specifically, an intronic SCN1A polymorphism was associated with short-term memory performance in four independent populations. Furthermore, fMRI experiments during a two-back working memory task revealed allele-dependent differences in brain activations in regions typically involved in working memory processes.
In the hypothesis-generating sample of 333 participants, we observed a medium effect size (ES) of the SCN1A SNP rs10930201 on short-term memory performance (Cohen's f=0.24). Recent candidate gene and genome-wide studies reporting associations of genetic polymorphisms with specific memory-related phenotypes observed also medium to large ES with Cohen's d-values ranging from 0.4 to 0.8 (for example, SNPs in BDNF,3 GRM328 and KIBRA5, 29). Therefore, the medium ES observed in this study is within the range of ES expected in behavioral genetics studies related to human memory capacity. Assuming a SNP with a medium ES (Cohen's d=0.5–0.6), the power of our initial sample to detect associations at a genome-wide FDR-corrected alpha error probability ranges from 67 to 89%. Thus, our initial sample is sufficient for detecting SNPs with medium to large ES, whereas it is underpowered for SNPs with small ES. The ES of the SCN1A SNP rs10930201 on brain activity as estimated in the fMRI study (Cohen's d=1.9) was considerably larger than the corresponding ES in the behavioral study. Such large ES are characteristic for imaging genetics studies of brain activity during memory tasks, as shown for example for several genes including BDNF (Cohen's d=2.2)3 and KIBRA (Cohen's d=1.6).5 A possible explanation for this repeatedly reported observation is that biological phenotypes such as neural activity are more proximate to the direct effects of genetic variability on gene products and their function, and might therefore be more sensitive in estimating genotype-dependent differences in mental processing.30, 31 Thus, the size of the effects of genetic variations on brain activity seems to be much larger as compared with behavioral measures. Consequently, in imaging genetics studies of memory, fewer participants are required to achieve enough statistical power to detect a significant genotype effect, than in behavioral genetics studies. Assuming a SNP with Cohen's d=2, the power of our fMRI sample to detect associations at an error probability equal to 0.001 is 85%. Thus, the fMRI sample is sufficient for detecting SNPs with ES typically observed in imaging genetics studies of brain activity during memory tasks.
It is important to stress that the intronic SCN1A SNP reported herein is most probably a marker SNP in linkage disequilibrium with one or more functional variations located elsewhere within SCN1A. As for any genetic association study, especially for GWAS on complex traits, the core message is the identification of a gene, rather than a specific variation, related to the phenotype of interest. In our case, additional issues require a cautious interpretation regarding the identification of the putative causative variants. First, SCN1A is essentially covered by one haplotype block, that is, it is hardly possible to exclude effects of neighboring and tightly linked SNPs. Second, SCN1A undergoes considerable alternative splicing,32 thereby showing the putative functional role of intronic, in addition to exonic, variants of the voltage-gated sodium channel Na(v)1.1. Database search (dbSNP) revealed that SCN1A harbors at least 481 tightly linked variations located in exonic, intronic and regulatory regions. One intronic SNP (rs3812718) located in the splice-donor site of SCN1A exon 5N, in the vicinity of our intronic SNP (5 kb apart) and in tight linkage disequilibrium with it (D′ and r2>0.9) was recently shown to have a striking effect on the percentage of transcripts of the neonatal form of the SCN1A gene in the adult brain.33 In those individuals with the common GG genotype, up to 50% of the transcripts include the neonatal version of exon 5, compared with an often undetectable level of the neonatal version in some participants with the rare AA genotype.33 Doses of both carbamazepine and phenytoin, two antiepileptic drugs targeting SCN1A, are highest in individuals with the AA genotype,34, 35 suggesting that the intronic SNP rs3812718 directly influences the therapeutic dose of antiepileptic medications and that it is possibly related to the influence of different SCN1A transcript variants on electrical signaling.33 To further study the relation of SCN1A variations on messenger RNA expression levels, we examined the association between SCN1A SNPs and cortical expression of the SCN1A transcript GI_29893558 in the brains of 178 non-demented deceased individuals (see Materials and Methods section). The data are based on the recently published genetic survey of human cortical gene expression.15 A subset of the SCN1A SNPs reported in our genetic study of human memory performance are also represented on the 500 k SNP Array set, which was used in the study of cortical gene expression. SNP rs4667869, which is located in the first SCN1A intron 8 kb 5′ to marker SNP rs10930201 and which is in linkage disequilibrium with the SNPs and the haplotype reported herein, is significantly and additively associated with messenger RNA expression of the SCN1A transcript (Supplementary Figure 3). In addition to these intronic SNPs, SCN1A also harbors a common non-synonymous SNP (rs2298771), which results in an amino acid substitution (Thr/Ala) in exon 16. Its functionality, however, is hitherto unknown. According to the HapMap database (http://hapmap.ncbi.nlm.nih.gov/), the intronic SNP rs10930201 and the non-synonymous SNP rs2298771 are in almost complete linkage disequilibrium (D′=0.96, r2=0.92), which was also confirmed in the 587 study participants from phases 1 and 2. Taken together, the SCN1A locus, which is associated with human memory performance, harbors multiple tightly linked variations with documented or putative functionality.
Voltage-gated sodium channels have an important role in the generation and propagation of the action potential and consist of an alpha subunit, which forms the ion conduction pore, and two auxiliary beta subunits.36 The alpha subunit has four homologous domains and different genes (SCN1A through SCN11A) encode different alpha subunits named Nav1.1 through Nav1.9.37 The SCN1A gene is located on chromosome 2q24 and contains 27 exons, spanning ∼139 kb of genomic sequence.38 SCN1A is expressed in brain regions critical for memory formation,39 regulates excitability of neuronal membranes and several SCN1A mutations are known to cause a variety of neurological diseases such as familial hemiplegic migraine,40 generalized epilepsy with febrile seizures plus type 241 and severe myoclonic epilepsy of infancy.42 Some antiepileptic drugs, such as phenytoin and carbamazepine, bind to voltage-gated sodium channels43 and genetic variability within SCN1A may predict the response to carbamazepine and phenytoin in patients diagnosed with epilepsy.34, 35, 44
Interestingly, phenytoin and carbamazepine have been shown to modulate working memory performance in rats. Lamotrigine, another antiepileptic drug that binds to voltage-gated sodium channels, is an effective maintenance treatment for bipolar disorder, particularly for prophylaxis of depression,45 a mental disorder with commonly observed working memory deficits.46 A recent fMRI study reports that lamotrigine treatment in depressed patients results in increased activation of brain regions typically involved in working memory processes during an n-back task,47 as also shown herein.
Taken together, a high-density GWAS followed by replication studies and an fMRI experiment and expression data suggest a role for the voltage-gated sodium channel Na(v)1.1 in human short-term memory. These results show the usefulness of unbiased genome-wide approaches and their potential to identify important molecular pathways related to human short-term memory. Ultimately, such studies might promote the development of targeted treatments of diseases associated with memory disturbances.
References
de Quervain DJ, Henke K, Aerni A, Coluccia D, Wollmer MA, Hock C et al. A functional genetic variation of the 5-HT2a receptor affects human memory. Nat Neurosci 2003; 6: 1141–1142.
de Quervain DJ, Papassotiropoulos A . Identification of a genetic cluster influencing memory performance and hippocampal activity in humans. Proc Natl Acad Sci USA 2006; 103: 4270–4274.
Egan MF, Kojima M, Callicott JH, Goldberg TE, Kolachana BS, Bertolino A et al. The BDNF val66met polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal function. Cell 2003; 112: 257–269.
Green AE, Munafo MR, Deyoung CG, Fossella JA, Fan J, Gray JR . Using genetic data in cognitive neuroscience: from growing pains to genuine insights. Nat Rev Neurosci 2008; 9: 710–720.
Papassotiropoulos A, Stephan DA, Huentelman MJ, Hoerndli FJ, Craig DW, Pearson JV et al. Common Kibra alleles are associated with human memory performance. Science 2006; 314: 475–478.
Mondadori CR, de Quervain DJ, Buchmann A, Mustovic H, Wollmer MA, Schmidt CF et al. Better memory and neural efficiency in young apolipoprotein E epsilon4 carriers. Cereb Cortex 2007; 17: 1934–1947.
Jessen F, Wiese B, Cvetanovska G, Fuchs A, Kaduszkiewicz H, Kolsch H et al. Patterns of subjective memory impairment in the elderly: association with memory performance. Psychol Med 2007; 37: 1753–1762.
Luck T, Riedel-Heller SG, Kaduszkiewicz H, Bickel H, Jessen F, Pentzek M et al. Mild cognitive impairment in general practice: age-specific prevalence and correlate results from the German study on ageing, cognition and dementia in primary care patients (AgeCoDe). Dement Geriatr Cogn Disord 2007; 24: 307–316.
Welsh KA, Butters N, Mohs RC, Beekly D, Edland S, Fillenbaum G et al. The Consortium to Establish a Registry for Alzheimer's Disease (CERAD). Part V. A normative study of the neuropsychological battery. Neurology 1994; 44: 609–614.
Wechsler D . Wechsler Memory Scale-Revised Manual. Psychological Corp: New York, NY, 1987.
Schmidt CF, Degonda N, Luechinger R, Henke K, Boesiger P . Sensitivity-encoded (SENSE) echo planar fMRI at 3 T in the medial temporal lobe. Neuroimage 2005; 25: 625–641.
Friston KJ, Holmes AP, Poline JB, Grasby PJ, Williams SC, Frackowiak RS et al. Analysis of fMRI time-series revisited. Neuroimage 1995; 2: 45–53.
Ashburner J, Friston KJ . Voxel-based morphometry—the methods. Neuroimage 2000; 11 (6 Part 1): 805–821.
Good CD, Johnsrude IS, Ashburner J, Henson RN, Friston KJ, Frackowiak RS . A voxel-based morphometric study of ageing in 465 normal adult human brains. Neuroimage 2001; 14 (1 Part 1): 21–36.
Myers AJ, Gibbs JR, Webster JA, Rohrer K, Zhao A, Marlowe L et al. A survey of genetic human cortical gene expression. Nat Genet 2007; 39: 1494–1499.
Baddeley A . Working memory. Science 1992; 255: 556–559.
Baddeley A . Working memory: looking back and looking forward. Nat Rev Neurosci 2003; 4: 829–839.
Owen AM, McMillan KM, Laird AR, Bullmore E . N-back working memory paradigm: a meta-analysis of normative functional neuroimaging studies. Hum Brain Mapp 2005; 25: 46–59.
Hautzel H, Mottaghy FM, Schmidt D, Zemb M, Shah NJ, Muller-Gartner HW et al. Topographic segregation and convergence of verbal, object, shape and spatial working memory in humans. Neurosci Lett 2002; 323: 156–160.
Wager TD, Smith EE . Neuroimaging studies of working memory: a meta-analysis. Cogn Affect Behav Neurosci 2003; 3: 255–274.
Braver TS, Cohen JD, Nystrom LE, Jonides J, Smith EE, Noll DC . A parametric study of prefrontal cortex involvement in human working memory. Neuroimage 1997; 5: 49–62.
Cohen JD, Perlstein WM, Braver TS, Nystrom LE, Noll DC, Jonides J et al. Temporal dynamics of brain activation during a working memory task. Nature 1997; 386: 604–608.
Jonides J, Schumacher EH, Smith EE, Lauber E, Awh E, Minoshima S et al. Verbal working memory load affects regional brain activation as measured by PET. J Cogn Neurosci 1997; 9: 462–475.
Schumacher EH, Lauber E, Awh E, Jonides J, Smith EE, Koeppe RA . PET evidence for an amodal verbal working memory system. Neuroimage 1996; 3: 79–88.
Jaeggi SM, Buschkuehl M, Etienne A, Ozdoba C, Perrig WJ, Nirkko AC . On how high performers keep cool brains in situations of cognitive overload. Cogn Affect Behav Neurosci 2007; 7: 75–89.
Rypma B, Berger JS, D'Esposito M . The influence of working-memory demand and subject performance on prefrontal cortical activity. J Cogn Neurosci 2002; 14: 721–731.
Rypma B, D'Esposito M . The roles of prefrontal brain regions in components of working memory: effects of memory load and individual differences. Proc Natl Acad Sci USA 1999; 96: 6558–6563.
Egan MF, Straub RE, Goldberg TE, Yakub I, Callicott JH, Hariri AR et al. Variation in GRM3 affects cognition, prefrontal glutamate, and risk for schizophrenia. Proc Natl Acad Sci USA 2004; 101: 12604–12609.
Schaper K, Kolsch H, Popp J, Wagner M, Jessen F . KIBRA gene variants are associated with episodic memory in healthy elderly. Neurobiol Aging 2008; 29: 1123–1125.
Hariri AR, Drabant EM, Weinberger DR . Imaging genetics: perspectives from studies of genetically driven variation in serotonin function and corticolimbic affective processing. Biol Psychiatry 2006; 59: 888–897.
Mattay VS, Goldberg TE, Sambataro F, Weinberger DR . Neurobiology of cognitive aging: insights from imaging genetics. Biol Psychol 2008; 79: 9–22.
Lossin C . A catalog of SCN1A variants. Brain Dev 2009; 31: 114–130.
Heinzen EL, Yoon W, Tate SK, Sen A, Wood NW, Sisodiya SM et al. Nova2 interacts with a cis-acting polymorphism to influence the proportions of drug-responsive splice variants of SCN1A. Am J Hum Genet 2007; 80: 876–883.
Tate SK, Depondt C, Sisodiya SM, Cavalleri GL, Schorge S, Soranzo N et al. Genetic predictors of the maximum doses patients receive during clinical use of the anti-epileptic drugs carbamazepine and phenytoin. Proc Natl Acad Sci USA 2005; 102: 5507–5512.
Tate SK, Singh R, Hung CC, Tai JJ, Depondt C, Cavalleri GL et al. A common polymorphism in the SCN1A gene associates with phenytoin serum levels at maintenance dose. Pharmacogenet Genomics 2006; 16: 721–726.
Catterall WA . From ionic currents to molecular mechanisms: the structure and function of voltage-gated sodium channels. Neuron 2000; 26: 13–25.
Plummer NW, Meisler MH . Evolution and diversity of mammalian sodium channel genes. Genomics 1999; 57: 323–331.
Wallace RH, Scheffer IE, Barnett S, Richards M, Dibbens L, Desai RR et al. Neuronal sodium-channel alpha1-subunit mutations in generalized epilepsy with febrile seizures plus. Am J Hum Genet 2001; 68: 859–865.
Yu FH, Mantegazza M, Westenbroek RE, Robbins CA, Kalume F, Burton KA et al. Reduced sodium current in GABAergic interneurons in a mouse model of severe myoclonic epilepsy in infancy. Nat Neurosci 2006; 9: 1142–1149.
Dichgans M, Freilinger T, Eckstein G, Babini E, Lorenz-Depiereux B, Biskup S et al. Mutation in the neuronal voltage-gated sodium channel SCN1A in familial hemiplegic migraine. Lancet 2005; 366: 371–377.
Escayg A, MacDonald BT, Meisler MH, Baulac S, Huberfeld G, An-Gourfinkel I et al. Mutations of SCN1A, encoding a neuronal sodium channel, in two families with GEFS+2. Nat Genet 2000; 24: 343–345.
Claes L, Del-Favero J, Ceulemans B, Lagae L, Van Broeckhoven C, De Jonghe P . De novo mutations in the sodium-channel gene SCN1A cause severe myoclonic epilepsy of infancy. Am J Hum Genet 2001; 68: 1327–1332.
Kuo CC . A common anticonvulsant binding site for phenytoin, carbamazepine, and lamotrigine in neuronal Na+ channels. Mol Pharmacol 1998; 54: 712–721.
Servin B, Stephens M . Imputation-based analysis of association studies: candidate regions and quantitative traits. PLoS Genet 2007; 3: e114.
Bowden CL, Calabrese JR, Sachs G, Yatham LN, Asghar SA, Hompland M et al. A placebo-controlled 18-month trial of lamotrigine and lithium maintenance treatment in recently manic or hypomanic patients with bipolar I disorder. Arch Gen Psychiatry 2003; 60: 392–400.
Robinson LJ, Thompson JM, Gallagher P, Goswami U, Young AH, Ferrier IN et al. A meta-analysis of cognitive deficits in euthymic patients with bipolar disorder. J Affect Disord 2006; 93: 105–115.
Haldane M, Jogia J, Cobb A, Kozuch E, Kumari V, Frangou S . Changes in brain activation during working memory and facial recognition tasks in patients with bipolar disorder with Lamotrigine monotherapy. Eur Neuropsychopharmacol 2008; 18: 48–54.
Acknowledgements
We thank Dr Heike Kölsch for expert support. This work was funded by the Swiss National Science Foundation (grants PP00B-106708, PP00B-68859, PP00B-114813, Sinergia, 3100-067114 to DQ, AP, and KH), by the German Research Network on Dementia (KND) and the German Research Network on Degenerative Dementia (KNDD), German Federal Ministry of Education and Research grants 01GI0420 and 01GI0711, and by the Ministry of Science, Republic of Serbia (project no. 145057Đ).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Molecular Psychiatry website
Supplementary information
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Papassotiropoulos, A., Henke, K., Stefanova, E. et al. A genome-wide survey of human short-term memory. Mol Psychiatry 16, 184–192 (2011). https://doi.org/10.1038/mp.2009.133
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/mp.2009.133
Keywords
This article is cited by
-
Heritability of human visual contour integration—an integrated genomic study
European Journal of Human Genetics (2019)
-
Multi-level genomic analyses suggest new genetic variants involved in human memory
European Journal of Human Genetics (2018)
-
Das Kompetenznetz Demenzen
Bundesgesundheitsblatt - Gesundheitsforschung - Gesundheitsschutz (2016)
-
Bridging Integrator 1 (BIN1) Genotype Effects on Working Memory, Hippocampal Volume, and Functional Connectivity in Young Healthy Individuals
Neuropsychopharmacology (2015)
-
Genome-Wide Analyses of Working-Memory Ability: A Review
Current Behavioral Neuroscience Reports (2014)