In the past decade, a group of laboratories from the School of Life Sciences, Nanjing University (SLiS of NJU) led by microRNA (miRNA) researcher Dr Chen-Yu Zhang, have focused on research in the field of extracellular miRNA (Fig. 1). The group’s original 2008 publication1 on the stable existence of serum miRNA is one of the most cited papers published by Chinese scholars in the past century and is the foundation for all serum miRNA biomarker studies. The work on secreted miRNA has led to extracellular vesicle-based delivery of small interfering RNA (siRNA) therapy. This white paper describes research undertaken by groups at SLiS of NJU, which includes a suggestion that plant miRNA could enter human circulation and exert biological functions, a new theory relating to traditional Chinese medicine (TCM), and a genetically engineered lettuce that could provide a therapy for the hepatitis B virus. We also describe work at SLiS of NJU on plant miRNA in pollen that determines whether honeybee larvae develop into a queen or worker bee, therefore deciphering the mystery of the honeybee caste formation. The multi-disciplinary group effort led to the theory that extracellular miRNA mediates the co-evolution between species.
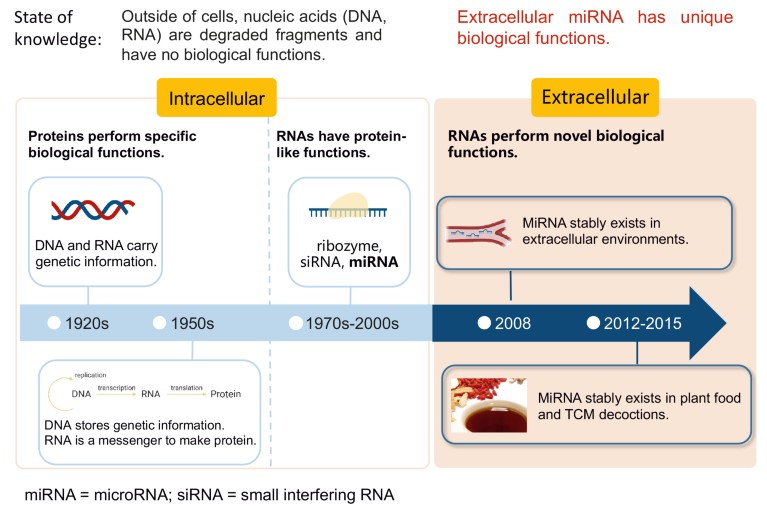
Figure 1. A three-stage history of nucleic acid study. DNA and RNA were known to carry genetic information but were thought to lack any further biological function. The discovery of ribozymes, small-interfering RNA and microRNA (miRNA) indicated that RNA can have protein-like functions, but it was still thought RNA was confined to the intracellular environment. In 2008, Dr Chen-Yu Zhang’s group discovered stable extracellular miRNA in blood1, and later also in plant food7 and TCM decoctions11. These findings indicate that miRNAs have a biological function outside the cell.
Serum microRNAs as potential biomarkers
MiRNAs are a class of single-stranded, small non-coding RNAs involved in post-transcriptional gene regulation. In 2008, Chen-Yu Zhang’s group demonstrated that serum miRNAs are stable in humans and animals1. Using deep sequencing, the group characterised the miRNA profile in human sera and identified unique patterns of serum miRNAs that distinguish patients with cancer and diabetes from healthy subjects. The findings suggest that the specific serum miRNA profiles may serve as fingerprints for disease diagnosis. In the same year, a separate study2 identified plasma miRNAs as promising biomarkers for prostate cancer. The findings challenge the convention that RNA is unstable in extracellular environments containing ribonucleases that degrade RNA. Building on other studies, accumulating evidence suggests that miRNAs circulate in a stable, cell-free form in extracellular biofluids including saliva, urine and breast milk and – because the extracellular miRNAs are tightly correlated with diseases such as cancers and diabetes – have potential to be diagnostic biomarkers3.
Secreted microRNA: Autocrine, paracrine or endocrine?
The finding that extracellular miRNAs circulate in body fluids, despite the presence of ribonuclease, indicates that mechanisms exist to protect miRNAs from degradation. Work at SLiS of NJU looked into whether extracellular vesicles (EVs) could function as a barrier to shield miRNAs from degradation. EVs, including nanometre-size exosomes and submicron-size microvesicles, are membrane-enclosed vesicular compartments that transport proteins, lipids and miRNAs in the extracellular environment. Zhang’s group found that the majority of circulating miRNAs were present in exosomes and microvesicles in human plasma rather than EV-free plasma4. The group proposed two models5 to explain the stability of extracellular miRNAs: (i) the protection of miRNAs by the membrane structures of EVs; and (ii) the stabilization of miRNAs by the formation of protein-miRNA complexes such as Argonaute 2. Moreover, they found that the miRNA profiles of EVs differ significantly from that of parent cells, and that cells selectively package different miRNAs into EVs when responding to different stimuli4. Therefore, the packaging or sorting of miRNAs into exosomes may be controlled by a specific mechanism. The secreted miRNAs can be delivered into recipient cells, where exogenous miRNAs silence the target genes and trigger downstream signalling events4. However, EV-encapsulated secreted miRNAs are not the only form of extracellular miRNAs. Extracellular miRNAs are generated through three main routes: (i) active secretion in EVs in an energy-dependent, selective process; (ii) release from donor cells in association with RNA-binding proteins; and (iii) passive leakage from broken or damaged cells owing to tissue injury, cell apoptosis or necrosis. It remains under debate whether EV-free miRNAs can be taken up by cells and participate in particular biological processes.
Work by Zhang’s research group on secreted miRNA suggests a new mechanism of intercellular communication: miRNAs secreted by a cell can either bind to autocrine receptors of the same cell to induce a signal (autocrine), transmit a local signal between nearby cells (paracrine), or travel to distal cells to spread a signal (endocrine). Intercellular communications mediated by hormone–receptor and antigen–antibody take place between certain types of cell, using cell-surface receptors. Secreted miRNAs have the potential to influence every type of cell, to deliver many types of miRNAs with each miRNA targeting multiple genes to affect specific target cells. Research into the biological relevance of miRNAs has shown that abnormally secreted miRNAs can lead to dysfunction and disease6 (Fig. 2). Undoubtedly, the elucidation of this novel siRNA transfer system will herald a new era in our understanding of signal transfer between cells.
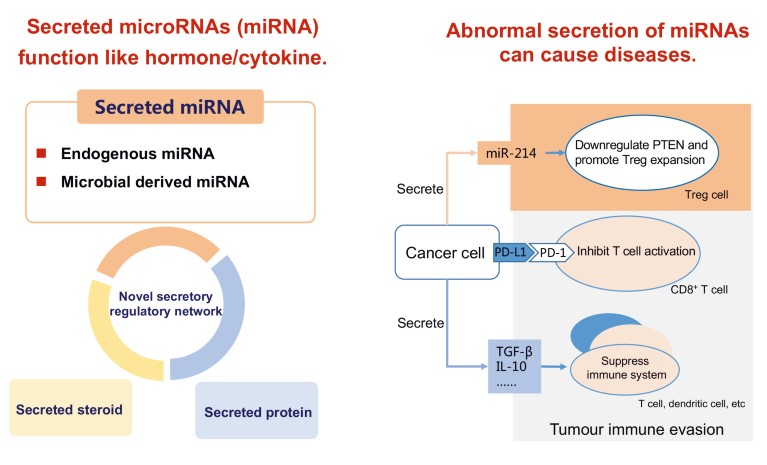
Figure 2. The microRNAs (miRNAs) secreted by endogenous cells circulate in body fluids and are delivered into recipient cells where they can silence the target genes and trigger downstream signalling events. Normal secretion of extracellular miRNAs maintains physiological homeostasis. Abnormal secretion can cause disease: for example, tumor cells can utilize two mechanisms to evade immune surveillance: i) upregulation of don’t find me signal on cell surface such as PD-L1 to evade the inspection by PD-1 on T cells; and ii) direct modulation of the host immune cells by secretion of anti-inflammatory cytokines such as TGF-β and IL-10. Dr Chen-Yu Zhang’s group provides the first evidence that tumor cells can actively modulate the host immune system via the secretion of miRNAs. Secreted miR-214 from tumour cells can promote the expansion of regulatory T cells (Tregs) and facilitate tumour immune escape through downregulation of PTEN in recipient CD4+ T cells6.
Plant microRNAs: We are what we eat
The focus of Zhang’s research group turned to whether miRNAs could transfer between distantly related, complex organisms and, if so, whether miRNAs could facilitate crosstalk between species. On the basis that humans consume plant-based foods such as grains, vegetables and fruits every day, the group hypothesized that exogenous plant miRNAs could withstand the acidic environment in the stomach and the digestive enzymes in the gastrointestinal tract and enter tissues.
Gene sequencing can determine whether miRNA is of plant or animal origin. Zhang’s bioinformatics group identified about 40 types of plant miRNAs after analysing serum RNA obtained from blood samples from healthy Chinese donors; some plant miRNAs were present at concentrations comparable to major endogenous human miRNAs7. Two plant miRNAs with the highest concentrations, MIR156a and MIR168a, which have been reported in rice (Oryza sativa) and crucifers (Brassicaceae), were acquired through food intake7. Research demonstrated that MIR168a could bind to low-density lipoprotein receptor adapter protein 1 (LDLRAP1) miRNA, inhibit LDLRAP1 expression in mouse liver and consequently regulate low-density lipoprotein removal from plasma7. The study indicates a possible model of cross-kingdom miRNA transfer in which plant miRNAs can survive gastro-intestinal digestion in mammals and when inside endogenous cells can elicit functions by regulating target genes and influencing physiological conditions.
‘You are what you eat’ is a proverbial saying in many countries. On the basis that foods confer health, Zhang’s research group is studying the biological mechanisms behind the benefits that eating plants can bring to humans. Diverse plant foods may bear distinct information in the form of different miRNA type and concentration, thereby generating different forms of gene regulation patterns and physiological impact on consumers. Therefore, we are not only eating food, but also eating “signals” and “information”. By studying cross-kingdom miRNA transfer they hope to find out whether dietary miRNAs have nutritional value. Important nutrients in foods include carbohydrates, proteins, lipids, vitamins and minerals. Plant miRNAs may represent a previously uncharacterized but essential bioactive compound in food. For instance, because regular consumption of plants is recommended to lower the risk of cardiometabolic diseases, cancers and age-related functional disorders, Zhang’s group asked whether dietary plant miRNA plays a part in preventing disease.
The existence of plant miRNAs in the tissues of animal models including pig, panda and rodent; the biological relevance of plant miRNAs in the prevention and treatment of human diseases have also been exhibited, including cardiovascular diseases, tumours, chronic inflammation and pulmonary fibrosis8. Cross-kingdom miRNA transfer may be a conserved and universal phenomenon that involves many complex organisms. With the rapid development of this field, more cross-kingdom miRNAs will be discovered along with their biological effects. Future studies should explore how miRNAs move across the boundary of different kingdoms, i.e., how plant miRNAs pass through the gastrointestinal tract and are absorbed by mammalian cells.
Cross-kingdom regulation and microRNA, to “bee” or not to be, that’s the question
From an evolutionary point of view, cross-kingdom miRNA transfer holds a unique position in facilitating cross-talk, communication and signal spreading in distant species from different kingdoms. This mechanism may shed new light on our understanding of the evolution of many extant species and provide new insight into their interactions and inter-dependence. Xi Chen’s group from SLiS of NJU sought to broaden their understanding of miRNA transfer between species and selected the honeybee (Apis mellifera) as a model organism to study. Female honeybees live in a social system and are divided into two castes: queens and workers. Queens are reproductive, have a larger body size, develop faster and live for longer than worker bees. Worker bees are sterile and are destined for a lifetime of intensive tasks such as housekeeping and food collection. Honeybee larvae are genetically identical but the fate of the larvae depends upon their diet. Larvae fed a diet of rich royal jelly develop into queens, whereas larvae that eat a less refined diet of ‘beebread’ and pollen develop into worker bees. Since beebread and pollen are mainly plant-originated, while royal jelly is a glandular secretion of nurse bees, the diets for worker- and queen-destined larvae are differentially derived from plant- and animal-sources, respectively. Chen hypothesized that the origin of the RNA in larval diets may impact honeybee development. To test the hypothesis, Chen’s research group measured miRNA levels in royal jelly, honey, beebread and pollen and found that beebread and pollen had much higher concentrations of plant miRNAs than did royal jelly9. The team suggests that supplementing royal jelly with plant miRNAs from pollen is one factor that prevents larvae from becoming a queen. Mechanistic studies suggest that Apis mellifera TOR (amTOR), a stimulatory gene in caste differentiation, is inhibited, at least in part, by plant MIR162a, helping to prevent larval differentiation into queens and inducing development towards worker bee phenotype9. The study suggests a mechanism for honeybee caste formation and also a system in which extracellular miRNAs could be part of cross-kingdom regulation.
The reason why honeybees use such a sophisticated and intricate mechanism to regulate the queen–worker dimorphism is a puzzle. Because only one single reproductive female queen is allowed, the caste structure has to be tightly regulated. In Chen’s theory, the reliance upon beebread and pollen as the exclusive food for larvae destined to become sterile workers may have evolved in concert with the exploitation of plant miRNAs for caste regulation through a form of ‘RNA interference (RNAi) castration’. Given that the effect of a larval diet that contains beebread is to help maintain the stability of the colony’s social order, it seems that plant miRNAs are not accidentally included in the larval diet but are collected with purpose, possibly to ensure the survival of the colony. The cross-kingdom regulatory function of plant miRNAs may have evolved with the selection of food sources by honeybees. Flowering plants have developed the characteristics that are attractive to honeybees for pollination, and the regulatory effects of plant miRNAs on caste differentiation in honeybees seems, at least in part, to influence the characteristics of the pollinators (Fig. 3). The selection of food sources by honeybees indicates an extraordinary evolutionary adaptation for colony success through partnership between two interacting organisms. Thus, honeybees and flowering plants exert selective pressures on each other in a co-evolutionary relationship, thereby affecting each other’s destiny in the inter-connected ecosystem.
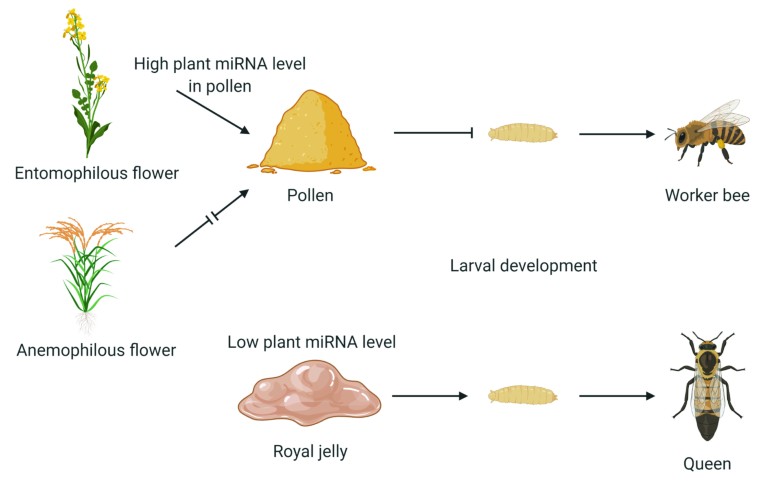
Figure 3. Entomophilous plants are insect-pollinated and contain microRNAs (miRNAs) that influence the size, morphology, colour and development of flowers and produce pollen that is attractive to insects including honeybees. Bee larvae that are fed pollen develop into worker bees because they receive a higher quantity of plant miRNAs than the larvae that receive only royal jelly9.
Is this kind of “borrowing” RNA substance from another species to regulate the process of their own survival and adaptation a common phenomenon? Considering that species have experienced millions of years of co-evolution to form the ecosystem we live in today, it is possible that cross-kingdom miRNA transfer may be frequently seen in nature. Indeed, emerging evidence has revealed that miRNAs can travel across the boundaries of species or kingdoms to spread gene silencing signals, thereby serving as a bond connecting the animal, plant and microbial kingdoms10. Such a phenomenon may make the study of miRNAs fundamental to shed light on the biological relevance of these molecules at an ecological and evolutionary scale.
miRNA from herbal medicine, plants vs. zombies?
TCM, especially herbal medicine, has been used in China to treat and prevent disease for thousands of years. However, many of the theories and practices employed in TCM are scientifically unsubstantiated because they have not been rigorously tested in randomized, controlled clinical trials. Some researchers have attributed the effects of TCM to the content of secondary metabolites and small molecules; macromolecules, such as RNA, have not been considered. RNA is thought to be unstable in the gastrointestinal tract and, because TCM herbs are usually boiled in water for several hours to produce a decoction, the assumption is that RNA is degraded during the preparation process. Consequently, scientific research on TCM decoctions often excludes RNAs from the biologically active ingredients.
The suggestion that plant-derived small RNAs (miRNA is classed as a small RNA) may affect human physiology has led to a new avenue of research in TCM. Some ultra-stable extracellular miRNAs retained in TCM herbs and decoctions for the prevention and treatment of diseases in TCM clinical practice are being studied. One example is honeysuckle (Lonicera japonica), a well-known Chinese plant used in TCM to treat infections with influenza virus. Zhang’s group found that MIR2911, a honeysuckle-encoded atypical miRNA, was selectively retained in the boiled honeysuckle decoction and had excellent stability and an intact sequence11. The group suggests that the high guanine and cytosine (GC) nucleotide content and unique sequence of MIR2911 may lead to its stability during the boiling process because two mutant versions of MIR2911 with reduced GC content were not stable. In vitro study11 suggests that MIR2911 could target and counteract multiple viral genes of influenza A viruses (IAVs). In vivo evidence demonstrates that gavage feeding the honeysuckle decoction leads to a significant elevation of MIR2911 in mouse blood and lung. Also, MIR2911 inhibited the replication in mouse models of various IAVs, including H1N1, H5N1 and H7N9, and alleviated viral infection-induced weight loss and reduced mortality. The study provided the first evidence that plant miRNA could be an active component in TCM and identified the first plant miRNA that could help to suppress viral infection (Fig. 4).
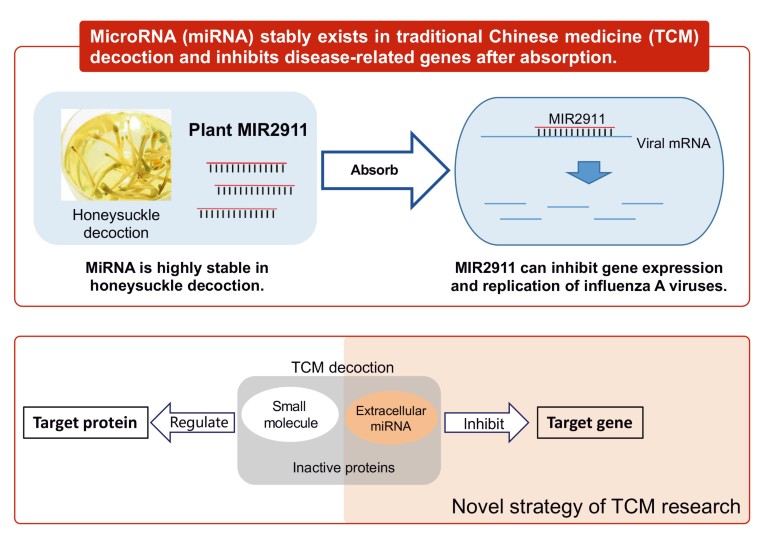
Figure 4. Some highly stable microRNAs (miRNAs) have been found in traditional Chinese medicine honeysuckle (Lonicera japonica) decoctions. Research suggests that an atypical miRNA called MIR2911, identified in honeysuckle decoction, binds to viral mRNA and exerts an inhibitory effect on the replication of influenza A viruses11.
The mechanisms through which miRNAs resist degradation when boiled with water to prepare TCM decoctions are not yet understood. Studies by the Chengyu Jiang research group in Peking Union Medical College, Beijing, may provide an answer. The group suggests that the heating process facilitates co-assembly of small RNAs and lipids in the decoctions to reach a steady state. The resulting heat-stable exosome-like nanoparticles, called ‘decoctosomes’, could enter human cells to deliver small RNAs that may exhibit therapeutic effects in vivo12. The results suggest a route through which lipids form liposomes with small RNAs in boiling decoctions to facilitate the uptake of small RNAs into human cells.
The SARS-CoV-2 pandemic has highlighted the threat that viruses can pose to public health. Viruses can rapidly mutate, which means that developing effective vaccines can be a challenge. Developing broad-spectrum antiviral drugs can help to prepare for future disease outbreaks. An MIR2911 and MIR2911-enriched honeysuckle decoction may be a therapeutic strategy to subdue viral infection. Besides targeting various subtypes of IAVs, studies suggest that honeysuckle-derived MIR2911 could directly inhibit the replication of varicella-zoster virus (the pathogen that causes chicken pox on primary infection and herpes zoster or shingles on reactivation) and enterovirus 71 (a primary pathogen of hand, foot and mouth disease in children) by targeting viral genes13,14. The studies indicate broad-spectrum antiviral activity by MIR2911. Therefore, a MIR2911 and MIR2911-containing honeysuckle decoction could be a potential therapeutic treatment for people infected with various forms of viral infections.
From science to biotechnology, a booming industry
The ultimate goal of fundamental research is to translate new discoveries into real-world applications. From this perspective, the presence of miRNAs in the extracellular environment has served as a stimulus to three important avenues of research: i) the potential that extracellular miRNAs may serve as disease biomarkers; ii) the potential of EV-encapsulated miRNAs to serve as a novel mode of RNAi-based gene therapy; and iii) the potential to engineer edible plants to biosynthesize and deliver miRNA medication for use in oral gene therapy.
The ability to diagnose a disease at an early stage is clearly beneficial. Diagnostic techniques such as biopsy, endoscopy and laparoscopy are usually invasive, but a blood sample could be a more efficient tool to help researchers identify biomarkers in serum or plasma. An ideal blood biomarker would fulfil many criteria, including: a high degree of specificity and sensitivity; enabling clinicians to detect a disease in its early stages and monitor a patient’s physiological and pathological status; remaining stable within a sample; and allowing rapid and accurate detection. Circulating miRNAs meet most of the criteria. For complicated diseases such as cancer, the variability between patients means that using just one protein marker is an unreliable method to determine disease status. In contrast, fingerprints consisting of a set of circulating miRNAs would have higher sensitivity and specificity compared to protein markers. Chen-Yu Zhang’s group has revealed that circulating miRNAs are useful in screening and detection in the early stages of some diseases15, which may be because cells can spontaneously secrete increased amounts of miRNAs into the blood in response to the early onset of diseases. Circulating miRNAs are stably present in blood and can have high abundance and integrity, even after a long period of sample storage. The concentration of circulating miRNAs is relatively low (that is, in the femtomolar range), but by using tools such as quantitative reverse transcription-polymerase chain reaction, microarrays and RNA sequencing they can be detected in a small sized sample. One of the bottlenecks in the field is the lack of standardized methodology to assess circulating miRNAs. Future approaches to help identify miRNAs as potential diagnostic tools could be to develop methods to stratify miRNA-containing EVs and miRNA-bound protein complexes, and to research sensitive techniques to identify EVs based on tissue origin and surface proteins.
RNAi therapeutics
Chen-Yu Zhang’s group from SLiS of NJU has explored the therapeutic potential of EVs as delivery vehicles for small RNAs. The group is interested in RNAi therapeutics but the lack of a safe and efficient delivery system has hindered its clinical application. Zhang’s group reasoned that they could recruit the body’s own RNA transport system – EVs. EVs are natural vesicles secreted by endogenous cells to protect and transport small RNAs across cells and biological barriers, therefore should be biocompatible with the host’s immune system. In several studies, Zhang’s group achieved potent target gene silencing and symptom relief in animal models16,17. Clinical applications of EV-mediated small RNA delivery will depend upon solving problems such as standardizing the complicated purification protocols of EVs, developing feasible methods to harvest sufficient quantities of EVs for clinical use, improving robust methods to load siRNA cargo into EVs and designing techniques to manipulate EV content and delivery. Research groups at SLiS of NJU envision that EV-mediated small RNA therapeutics will develop rapidly and become a breakthrough in gene therapy.
Despite significant advances in injectable, transdermal and nasal routes for drug administration, the oral route is preferred because of its convenience for the patient. Unfortunately, oral administration of RNAi therapeutics has been hampered by physiological barriers in the gastrointestinal tract. Advances in cross-kingdom miRNA transfer may help to overcome the major challenges. Plants seem to be ideal carriers for therapeutic miRNAs because some plant miRNAs are resistant to the digestive process. Zhi Hong’s group from SLiS of NJU have developed a strategy to deliver small RNAs as therapeutic medication by engineering transgenic plants to express therapeutic miRNAs. The group engineered a lettuce to biosynthesize artificial miRNAs to specifically target the hepatitis B virus surface antigen gene (HBsAg) with the endogenous miRNA biogenesis machinery of the lettuce18. After oral administration of the lettuce decoction, artificial miRNAs were absorbed and delivered into the liver to inhibit HBsAg expression in transgenic mice. After 15 months of treatment, the expression of HBsAg was reduced and liver injury was significantly alleviated in the mice and no toxicological effects were observed18. This strategy utilizes the plant endogenous miRNA biogenesis machinery to produce methylated miRNAs for increased stability while significantly reducing the cost of production. Plant miRNA-based oral therapy may provide an effective, nontoxic and affordable treatment option for disease.
To facilitate and accelerate the commercialization of these scientific breakthroughs, Nanjing University has established NJU Advanced Institute of Life Sciences; Jiangsu Engineering Research Center for MicroRNA Biology and Biotechnology; and Nanjing Drum Tower Hospital Center of Molecular Diagnostic and Therapy. Very recently, Nanjing University has established the Institute of Artificial Intelligence Biomedicine, a big data-empowered tech transfer platform that aims to connect all the dots from science to technology to products. The future goal is that the original scientific theories created by Dr Chen-Yu Zhang’s ‘Nanjing school of thought’ will become medical innovations that could benefit the health of all people.


