In situ sequencing (ISS)
ISS is a technology that enables the analysis of hundreds of genes with sub-cellular resolution, at their original location in morphologically preserved tissue sections. This is achieved by generating and sequencing, directly inside these sections, clonally amplified barcode sequences that are introduced by ligation of gene specific probes. Key advantages of ISS over alternative multiplexed in situ hybridization or spatial sequencing methods are its high specificity and throughput (~15 cm2 of tissue area per week per microscope).
CARTANA (www.cartana.se) has made ISS commercially available by providing sample preparation kits with customizable gene panels (Fig. 1a), and ISS kits with optimized decoding chemistry. The sample preparation step generates 0.5–1 μm barcoded DNA spots at the original location of a detected RNA molecule (Fig. 1b–d). These DNA spots are then sequenced directly inside the tissue section, using the ISS kit and an epifluorescence microscope. During the ISS reaction cycles, fluorescently labeled probes are iteratively attached, imaged and removed, making it possible to read the barcode sequences in the DNA spots (Fig. 1e-g). Key advantage of the CARTANA ISS chemistry is the high intensity and high signal-to-noise ratio that enables imaging with low magnification objectives. Hence, ISS reactions of large tissue areas are imaged with high speed, significantly increasing throughput in comparison to other in situ transcriptome analysis techniques.
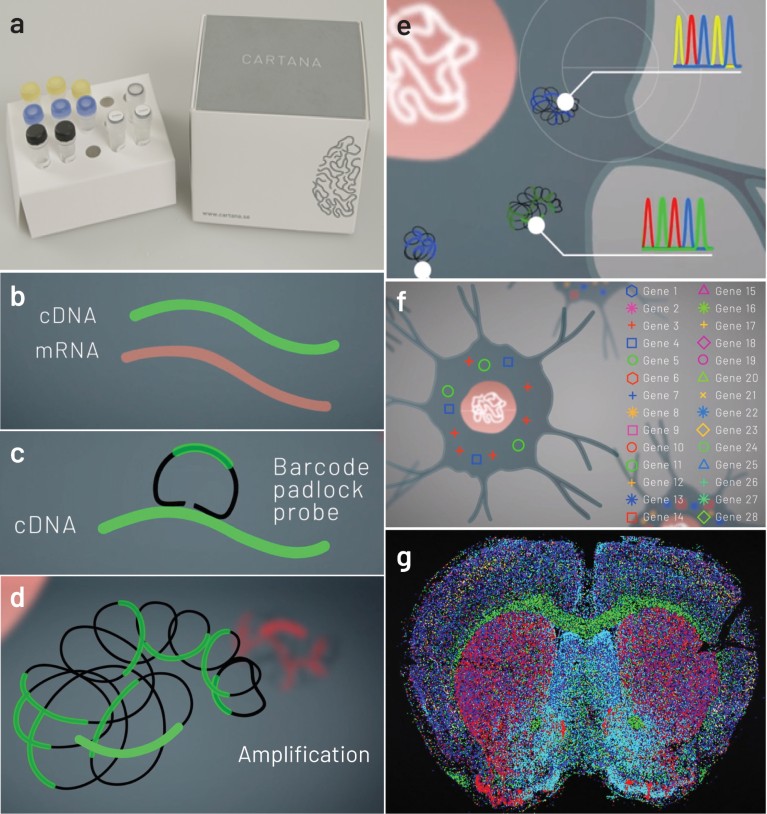
Figure 1 | CARTANA ISS technology for high throughput spatial transcriptomic analysis. a, CARTANA sample preparation reagent kit. b, mRNAs of cell-type markers on preserved tissues are fixed and reverse transcribed to cDNA. c, Barcoded probes are hybridized and ligated onto the target cDNA. d, Ligated probes are locally amplified, resulting in DNA spots containing multiple copies of the barcodes. e, Barcodes are sequenced in the tissue by five cycles of ISS chemistry and fluorescence imaging. f, After decoding the barcode sequences, the detected gene marker identities are assigned to every DNA spot. g, The target genes are visualized in whole-tissue section.
Fluorescent imaging during ISS
The Nikon Ti2-E inverted microscope is particularly well-suited to high-throughput imaging of ISS reads. The Perfect Focus System 4 (PFS4) focus-locking system and integrated motorized stage provides the stability needed for performing high-throughput assays without interruption. Complex multidimensional acquisitions can be performed with confidence. This includes large area scanning, z-stacks and multiple fluorescence channels. The Ti2-E enableshigh-quality ISS within several whole-tissue sections, in up to nine slides in parallel, per experiment.
Applying ISS to quantify RNA molecules in single cells of a mouse brain section.
Fresh frozen 10 μm mouse brain sections are fixed and prepared for ISS using the CARTANA sample preparation kit, containing a customized probe panel targeting 150 central nervous system marker genes(Fig. 2a). The generated barcoded DNA spots are then sequenced with CARTANA ISS kit chemistry with high signal-to-noise ratio (Fig. 2b–g). Next, the identities of all DNA spots are decoded and the 150 targeted genes are spatially mapped with sub-cellular resolution (Fig. 2h-j).
The Ti2-E microscope with ×20 objective and sCMOS camera is used to image each ISS cycle of the brain tissue section (8.3 mm × 6.2 mm × 10 μm). In a pre-scan, the tissue area on the microscope glass slide is defined, the camera exposure optimized and other settings adjusted using Nikon’s NIS-Elements software.
Thereafter, automated image acquisition, consisting of 165 fields of view (10% overlap between individual fields) and 15 z-planes (0.8 μm steps), is performed for five fluorescence channels: DAPI to visualize the cell nuclei, and four channels for ISS chemistry. One ISS imaging cycle of this tissue dimension is completed in less than one hour. To decode the barcodes of the 150 targeted genes, five ISS cycles are required. An optional cycle for antibody staining can be added. After image acquisition, the NIS-Elements software performs maximum intensity projections, large image stitching and image export.
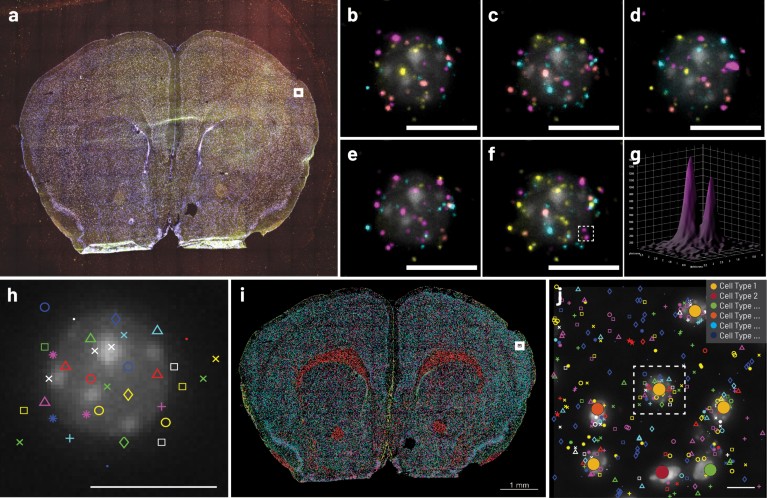
Figure 2 | Brain cell typing with CARTANA ISS. a, Raw, multicolor image of whole tissue section after sample preparation and one ISS cycle, imaged with a Nikon Ti2-E microscope. b–f, Close-up of the white square region in a, displaying a single cell undergoing five cycles of ISS. g, Surface plot displaying the fluorescence intensity (a.u.) of two sequenced DNA spots inside the dashed box in f. h, Barcodes are decoded to assign gene markers (pseudo-colored symbols) to each DNA spot. i, Generated cell-type map of the entire tissue section. Each cell is assigned to a cell-type based on the gene markers in proximity and unsupervised clustering. j, Zoom-in of white box in i, dashed box corresponds to the cell in h. Scale bars, 10 μm (b–f,h,j).
In situ cell-type classification by ISS
The consolidated analysis of each decoded DNA spot results in ahigh-density gene expression map (Fig. 2i) over the entire tissue area. The DAPI nucleus staining can be used to segment individual cells and assign ISS gene reads by spatial proximity (Fig. 2j). Due to itshigh-throughput, ISS is exceptionally well suited for mapping of cell types in tissue sections that were defined by single-cell RNA sequencing. Generating large spatial cell atlases enables the analysis of unique gene expression patterns and relationships of different cell types comprising tissues and their architecture.
Outlook
Sample preparation and ISS kits developed by CARTANA, together with the Nikon Ti2-E microscope system and NIS-Elements software, provide a powerful solution for quantitative spatial analysis of hundreds of genes with single-cell resolution, high-throughput and high specificity. This technology can contribute to scientific breakthroughs in many research areas, including neuroscience, oncology, immunology and embryonic development, generating new insights about cellular and molecular biology, and supporting the development of diagnostics and treatments.
ADDITIONAL INFORMATION
John R. Allen, at Nikon Instruments Inc, Melville, NY, USA, is a non-author contact at john.allen@nikon.com



 Nature Methods
Nature Methods