Scientists now have an unprecedented view of the genetic changes that can contribute to cancers, such as squamous cell carcinoma, a common form of mouth cancer.Credit: Steve Gschmeissner/SPL
This week, Nature is publishing a suite of papers that sheds new light on the genetic causes of cancer. The results show how far our understanding of cancer has come — and how far we still have to go.
The Pan-Cancer Analysis of Whole Genomes Consortium brought together researchers with nearly 750 affiliations across 4 continents. Between them, they sequenced full genomes from more than 2,600 samples representing 38 different types of cancer. The work is summarized in a News & Views article in this issue.
The project is remarkable in both scope and complexity, and, partly because of this, faced challenges at every step; from acquiring samples to protecting patient privacy while putting terabytes of data into the hands of researchers.
Global genomics project unravels cancer’s complexity at unprecedented scale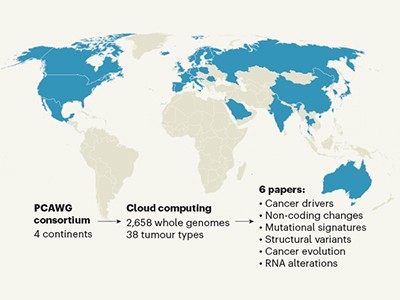
Thanks to these efforts — and previous full-genome sequences — scientists now have an unprecedented view of the genetic changes that can contribute to cancer, and a clearer idea of where gaps in knowledge remain. Altogether, the team pinpointed 705 mutations that occurred repeatedly in the cancer genomes, suggesting that they are important for tumour growth. Of these, about 100 fell outside the protein-coding regions of the genome, but more such mutations might be uncovered with improvements in computational techniques for analysing non-coding regions. Overall, the authors found that cancer genomes contain an average of four to five mutations that drive tumour growth. In 5% of cases, however, they found no such mutations.
Cancer genomes have been sequenced for more than a decade, but now researchers and the funders who support them must tackle the next challenge. The goal has always been to improve the lives of those affected by cancer, and the reams of data amassed by sequencing projects have helped. They are used by researchers to find new drug targets, and to generate new markers that can be used to match patients with the treatment most likely to help.
But most of the data so far have been limited in one crucial respect: clinical details of the sample donors are often missing. The first samples collected for the Cancer Genome Atlas, a sequencing project that ran from 2006 to 2018, co-funded by the US National Cancer Institute and the National Human Genome Research Institute, typically came with little more than the donor’s gender, diagnosis and age at diagnosis. Rarely would there be a record of that person’s family or medical history, what therapy they had received and how they had responded — all crucial information if genome sequences are to be put to work to help patients.
The next generation of cancer-genome sequencing projects is trying to change that. But gathering detailed clinical information is more difficult — and more expensive — than sequencing genomes, particularly in the many countries that lack a unified health-care system. There, accessing hospital records is complicated: different hospitals keep records differently; patients often move from one treatment centre to another; and the quality of records varies enormously. More-detailed records also mean greater risk of personal exposure if there is a privacy violation, raising the bar yet again for participant protection.
Genomics: data sharing needs an international code of conduct
These are all pressing issues, not only in cancer research, but in health care generally. Efforts are already under way to transform health records into a format that can be more readily, but securely, accessed and studied. The American Association for Cancer Research’s project GENIE, for example, has compiled 70,000 records of tumour DNA sequences, and real-world clinical data. The United Kingdom’s 100,000 Genomes Project also aims to match DNA sequences with clinical information for a variety of conditions. And the International Cancer Genome Consortium, which has coordinated much of the tumour sequencing work so far, has launched a new phase, this time with a focus on clinical information.
Pooling large numbers of samples is a powerful way to find genetic changes that can drive cancer, and provides a starting point for learning how they do so. But the real return on investment will come when that information can be used to tailor therapy to individual patients. And for that to be achieved, clinical background information on study participants is essential.
When cancer-genome sequencing projects were first launched, it was hoped that they would provide a catalogue of mutations that could give rise to cancer — and reveal broad patterns on which researchers could base drug development. The core of that mission has been achieved, but many cancers have proved more complex than expected. Seemingly similar cancers can contain very different sets of mutations — no two cancers are quite the same.
As is often the case in biomedical research, the answers to a question are more complex than originally imagined. But recognizing the complexity is empowering, and harnessing it will be necessary in the search for better treatments.

 Read the paper: Pan-cancer analysis of whole genomes
Read the paper: Pan-cancer analysis of whole genomes
 Global genomics project unravels cancer’s complexity at unprecedented scale
Global genomics project unravels cancer’s complexity at unprecedented scale
 Genomics: data sharing needs an international code of conduct
Genomics: data sharing needs an international code of conduct
 Faster, better, cheaper: the rise of CRISPR in disease detection
Faster, better, cheaper: the rise of CRISPR in disease detection
 Genome studies unlock childhood cancer clues
Genome studies unlock childhood cancer clues
 Nature Collection: Rare cancers
Nature Collection: Rare cancers