- NEWS AND VIEWS
Gene editing reveals the effect of thousands of variants in a key cancer gene
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Rent or buy this article
Prices vary by article type
from$1.95
to$39.95
Prices may be subject to local taxes which are calculated during checkout
Nature 562, 201-202 (2018)
doi: https://doi.org/10.1038/d41586-018-06022-y
References
Kuchenbaecker, K. B. et al. J. Am. Med. Assoc. 317, 2402–2416 (2017).
Plon, S. E. et al. Hum. Mutat. 29, 1282–1291 (2008).
Richards, S. et al. Genet. Med. 17, 405–424 (2015).
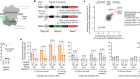
Findlay, G. M. et al. Nature 562, 217–222 (2018).
Blomen, V. A. et al. Science 350, 1092–1096 (2015).
Starita, L. M. et al. Am. J. Hum. Genet. https://doi.org/10.1016/j.ajhg.2018.07.016 (2018).
Rehm, H. L. et al. N. Engl. J. Med. 372, 2235–2242 (2015).
Spurdle, A. B. et al. Hum. Mutat. 33, 2–7 (2012).
Ma, L. et al. Proc. Natl Acad. Sci. USA 114, 11751–11756 (2017).
Rahman, N. Nature 505, 302–308 (2014).
Collins, F. S. & Varmus, H. N. Engl. J. Med. 372, 793–795 (2015).

 Read the paper: Accurate classification of BRCA1 variants with saturation genome editing
Read the paper: Accurate classification of BRCA1 variants with saturation genome editing
 Edit the genome to understand it
Edit the genome to understand it
 Complex assistance for DNA invasion
Complex assistance for DNA invasion