Abstract
MicroRNA (miRNA) biogenesis is finely controlled by complex layers of post-transcriptional regulators, including RNA-binding proteins (RBPs). Here, we show that an RBP, QKI5, activates the processing of primary miR-124-1 (pri-124-1) during erythropoiesis. QKI5 recognizes a distal QKI response element and recruits Microprocessor through interaction with DGCR8. Furthermore, the recruited Microprocessor is brought to pri-124-1 stem loops by a spatial RNA-RNA interaction between two complementary sequences. Thus, mutations disrupting their base-pairing affect the strength of QKI5 activation. When erythropoiesis proceeds, the concomitant decrease of QKI5 releases Microprocessor from pri-124-1 and reduces mature miR-124 levels to facilitate erythrocyte maturation. Mechanistically, miR-124 targets TAL1 and c-MYB, two transcription factors involved in normal erythropoiesis. Importantly, this QKI5-mediated regulation also gives rise to a unique miRNA signature, which is required for erythroid differentiation. Taken together, these results demonstrate the pivotal role of QKI5 in primary miRNA processing during erythropoiesis and provide new insights into how a distal element on primary transcripts affects miRNA biogenesis.
Similar content being viewed by others
Introduction
MicroRNAs (miRNAs) are versatile regulators of gene expression in complex organisms. Therefore, the findings that miRNAs themselves are tightly controlled to regulate gene expression in a temporally restrained and cell-specific manner, are not surprising1,2. The canonical pathway of miRNA biogenesis is initiated by the generation of primary miRNAs (pri-miRNAs), followed by two-step cleavage3,4. Two RNase III gene family members, Drosha and Dicer, are the key catalyzers in pri-miRNA cleavage. In particular, Drosha and its cofactor DGCR8/Pasha form a trimeric complex called Microprocessor that recognizes and cuts hairpin-containing pri-miRNAs to generate the intermediate precursor pre-miRNAs5,6,7. Once pre-miRNAs are exported to the cytoplasm, they are cleaved by Dicer, leaving two ∼22 nt strands that pair to each other with ∼2 nt 3′ overhangs8,9. One strand of each duplex (mature miRNA, hereafter as miRNA) is loaded onto an Argonaute protein to form the core of the silencing complex, and the other strand is often discarded10.
Pri-miRNA processing is a critical step in miRNA biogenesis because it not only defines the terminus of a miRNA but also determines miRNA abundance. A typical metazoan pri-miRNA consists of an ∼33 bp stem with an apical loop and basal single-stranded RNA (ssRNA) segments, which are thought to be critical for Microprocessor recognition and accurate processing11. Recently, the stoichiometry of Microprocessor was revealed: Drosha recognizes and measures the basal ssRNA-dsRNA junction, whereas DGCR8 interacts with the apical elements to ensure processing fidelity7. Other sequence elements are also reported to be involved in pri-miRNA processing; the CNNC motif, residing in the basal region, could be recognized by SRp20 (also called SRSF3) or DEAD-box RNA helicase p72 (also called DDX17) and promote human pri-miRNA processing12. Additional processing features in the apical loop also contribute to pri-miRNA recognition and processing. For example, heterogeneous nuclear ribonucleoprotein A1 (HNRNPA1) and KSRP bind to the apical loop of pri-miR-18a and pri-let-7, respectively, and facilitate Drosha-mediated processing13,14. LIN28 recognizes the apical loop of pri-let-7 and suppresses its processing by Drosha15,16,17. Nevertheless, including the UG motif in the basal region and the UGUG motif in the apical loop12,18, all these functional motifs only account for ∼80% of human miRNAs, suggesting that other determinants may exist and contribute to efficient recognition and processing by Microprocessor.
In contrast to previously reported cis-elements that are embedded in or near the pri-miRNA stem loops to affect Microprocessor activity4, here we identified an RNA motif that could act at a distance to promote pri-124-1 processing by Microprocessor. In this case, QKI5 recognized the QKI response element (QRE) ∼300 nt upstream of pri-124-1 stem loops. Once activated, QKI5 recruited Microprocessor via interaction with the double DRSM domain of DGCR8. As this recruitment was a few hundred bases apart from pri-124-1 stem loops, we proposed a spatial RNA-RNA interaction, which brings the QRE and stem loops into close proximity, thus allowing QKI5-recruited Microprocessor to reach pri-124-1 stem loops and to activate its processing. Moreover, QKI5 regulates a unique miRNA signature, with some of them being miR-124-like attenuators for erythropoiesis. Thus, QKI5 contributes to the post-transcriptional regulation of miR-124 and other miRNAs, conferring the exquisite miRNA balance necessary for erythroid development. Notably, RNA-binding proteins (RBPs) bound to separate sites on pri-miRNA transcripts could act at a distance to orchestrate miRNA biogenesis, revealing that pri-miRNAs may differentially benefit from the different motifs that they carry.
Results
The expression of pri-miRNAs and miRNAs during erythropoiesis
To investigate whether miRNA biogenesis is regulated at the post-transcriptional level during erythropoiesis, we first measured the expression of both the primary and mature transcripts of 16 candidate miRNAs that have been reported to participate in either erythroid differentiation or myeloid leukemogenesis (Supplementary information, Table S1). We surveyed RNA samples from primary cultured CD34+ hematopoietic stem/progenitor cells (HPCs) undergoing erythropoietin (EPO)-induced erythroid maturation (Figure 1A) and two erythroleukemia cell lines (K562 and HEL) undergoing erythroid differentiation by hemin treatment (Supplementary information, Figure S1A). Dramatic expression changes of individual miRNAs were observed: (1) consistent alteration in the levels of both pri-miRNAs and miRNAs, such as increases in miR-451, -23a, -15a and -17 or decreases in miR-191, -223, -126 and -433; (2) irregular changes in the levels of either pri-miRNAs or miRNAs, such as miR-125b, -155, -221 and -146a; and (3) reverse relationship between pri-miRNAs (increased) and miRNAs (decreased), such as miR-128, -129, -150 and -124, which indicates a post-transcriptional control of their expression (Figure 1B, Supplementary information, Figure S1C and Table S2).
Post-transcriptional control of pri-124-1 during erythroid differentiation. (A) May-Grunwald-Giemsa staining shows the morphology of HPCs in erythroid culture (E culture). Scale bar, 10 μm. (B) Heatmap representation of q-PCR results of pri-miRNAs and miRNAs in HPCs, K562 and HEL cells. Each value was normalized by its percentage to the sum of expression at all time points (e.g., for pri-124-1, the value of its 0 h expression = 0 h/(0 + 24 + 48 + 72 h)). (C) The calculated relative miRNA processing efficiency in HPCs. (D) q-PCR analysis of the relative abundance of three primary transcripts corresponding to three miR-124 loci in the human genome. The expression of each transcript was normalized to pri-124-1 using the ΔΔCq calculation method. (E) Northern blot analysis of pri-124-1, pre-124-1 and miR-124 transcripts in HPCs, K562 and HEL cells in E culture. The upper panel shows hybridization of pri-124-1 with DIG-labeled RNA probes; 18s rRNA was used as a loading control. The lower panel shows hybridization of pre-124-1 and miR-124 with isotope-labeled DNA probes; U6 snRNA was used as a loading control. (F) q-PCR results shows the relative expression levels of pri-124-1, pre-124-1 and miR-124 in HPCs. Arrows indicate the primer pairs used for amplification. (G) A schematic representation of the regulated processing pathway of miR-124-1 during erythropoiesis. Error bars reflect SEM from three biological replicates if not stated otherwise. Significance was determined by t-test with *P< 0.05; **P< 0.01; ***P< 0.001.
Attenuated pri-124-1 processing is coupled with erythropoiesis
Due to the significant changes in pri-124-1 and miR-124 levels during erythropoiesis (Figure 1C and Supplementary information, Figure S1B), we decided to further investigate its regulation. As human miR-124 is represented at three different genomic loci (miR-124-1 (8p23.1), miR-124-2 (8q12.3) and miR-124-3 (20q13.33)), the transcription efficiencies of the multiple loci were first evaluated in different erythroid cells. Fortunately, miR-124 was primarily expressed from the miR-124-1 locus in HPCs, K562 and HEL cells (Figure 1D). Therefore, the subsequent analysis focused only on miR-124-1. To further confirm the opposing expression patterns of pri-124-1 and miR-124, northern blot was performed and showed that pri-124-1 was readily detectable and increased over the course of erythroid differentiation in HPCs and both cell lines (Figure 1E). In contrast, the precursor (pre-124-1) and miR-124 displayed expression changes opposite to that of pri-124-1, with decreased levels throughout the differentiation time course (Figure 1E). Similar results were also observed from quantitative real-time PCR (q-PCR) analysis (Figure 1F and Supplementary information, Figure S1C). Although the miR-124 level was much lower in cell lines compared with HPCs (Figure 1E), which might be due to their cancerous origin, the discrepancy between pri-124-1 and pre-124-1 changes is still obvious. Therefore, these data indicate attenuated pri-124-1 processing at the Microprocessor step during erythropoiesis (Figure 1G).
RBP screening identifies several pri-124-1-associated proteins
As this regulation occurs at RNA levels, we speculated that an RBP might participate in this process. As the first step to investigate the possible mechanisms for this regulation, the full-length transcript of pri-124-1 must be obtained. Therefore, 5′ and 3′ RACE were conducted in K562 cells, and a specific 961-nt RNA transcript (Supplementary information, Figure S1D and S1E) was identified to be perfectly mapped to the miR-124-1 locus in human genome. RBPs usually recognize their targets either in a sequence- or secondary structure-specific manner. Due to the flexible endogenous RNA structures, it is more reasonable to predict functional RBPs by their binding sequence motifs. Therefore, we used the RBPDB database (http://rbpdb.ccbr.utoronto.ca/) to perform this bioinformatics prediction (Supplementary information, Table S3). This algorithm yielded ∼20 RBPs with one or more binding sites on pri-124-1 (Supplementary information, Table S3 and Figure S2A). To screen the functional RBPs that could modulate pri-124-1 processing, we examined both pri-124-1 and miR-124 levels in K562 cells upon the introduction of RBP-specific siRNAs (Supplementary information, Figure S2B and Figure 2A). We speculated that two classes of RBPs participate in this process: (1) negative RBPs that attenuate pri-124-1 processing as shown by a decrease in the level of pri-124-1 but an increase in miR-124 level in response to RBP knockdown and (2) positive RBPs that promote pri-124-1 processing as shown by an increase in pri-124-1 level but a decrease in miR-124 level when it was reduced (Figure 2B). Accordingly, these results indicated KSRP, ELAV1 and EIF4B as the negative regulators, whereas QKI5, SRSF10 and PABPC1 as the positive regulators for pri-124-1 processing (Figure 2A and 2B). Of these RBPs, QKI5 was the most attractive candidate for the following reasons: (1) QKI5 knockdown led to the biggest difference in pri-124-1 (∼5-fold increase) and miR-124 (∼3-fold decrease; Figure 2A); (2) a nuclear localization signal in the C-terminus of QKI5 allows its import into the nucleus, where Microprocessor-mediated cleavage occurs; and (3) QKI5 was downregulated in HPCs and K562 cells upon erythroid induction (Figure 2B), consistent with the changes of miR-124 (Figure 1E); (4) previous studies showed contradictory results regarding the effect of QKI isoforms on miRNA expression19,20.
QKI5 regulates pri-124-1 processing during erythropoiesis. (A) q-PCR of endogenous pri-124-1 and miR-124 in K562 cells after 48-h transfection with individual RBP siRNAs. RBP candidates with opposite effects on pri-124-1 and miR-124 are indicated in red and blue, respectively. (B) A schematic representation of RBPs classified as either repressing (negative RBPs) or promoting (positive RBPs) pri-124-1 processing. The right panel shows immunoblot results of selected RBPs in both K562 cells and HPCs in E culture. (C) q-PCR analysis of pri-124-1 and miR-124 in K562 cells transfected with pCMV6-QKI5 or si-QKI5 for 48 h. Lower panel: immunoblot showing the levels of QKI5 (or Flag-QKI5) in pCMV6-QKI5- and si-QKI5-transfected K562 cells. Two sets of primers (#1 and #2, sequences are shown in Supplementary information, Table S4) were used to amplify pri-124-1. (D) q-PCR analysis of pri-124-1 and miR-124 in HPCs transduced with lenti-si-QKI5 or lenti-GFP in E culture. Lower panel: immunoblot of QKI5 in HPCs transduced with lenti-si-QKI5 or lenti-GFP in E culture. Error bars reflect SEM from three biological replicates if not stated otherwise. Significance was determined by t-test with *P< 0.05; **P< 0.01; ***P< 0.001.
QKI5 affects pri-124-1 processing
To further validate the regulatory effect of QKI5 on pri-124-1 processing, we performed q-PCR to measure the levels of pri-124-1 and miR-124 in K562 and HEL cells upon QKI5 overexpression or knockdown. As expected, QKI5 overexpression reduced pri-124-1 but increased miR-124 in both cell lines (Figure 2C, Supplementary information, Figure S2C and S2D). In contrast, QKI5 knockdown resulted in increase in the level of pri-124-1 but decrease in miR-124 level (Figure 2C, Supplementary information, Figure S2C and S2D). miR-23a (without QREs on pri-23a∼27a∼24-2), which is irrelevant to this pathway, showed no changes in response to QKI5 overexpression or knockdown (Figure 2C, Supplementary information, Figure S2C and S2D).
Similar to the above findings observed in the K562 and HEL cells, QKI5 knockdown by lentiviral delivery of RNAi vectors augmented pri-124-1 accumulation but decreased miR-124 in HPCs (Figure 2D). Furthermore, a luciferase reporter construct containing the miR-124-1 gene promoter was not affected by QKI5, eliminating the possibility of transcriptional induction of pri-124-1 by QKI5 (Supplementary information, Figure S2E). Collectively, these observations indicate that QKI5 specifically activates pri-124-1 processing in erythroid cells.
The regulation of pri-124-1 by QKI5 is dependent on its binding to QRE
A distal QRE was found ∼300 nt upstream of the pri-124-1 stem loops (Figure 3A). As QKI5 has been shown to modulate various mRNA targets via binding to a specific QRE21,22, we investigated whether this interaction also accounts for its regulation on pri-124-1 processing. Earlier studies have reported that the substitution of valine 157 to glutamic acid (V157E) in the KH domain of QKI5 abolished its ability to bind to QREs23. Thus, we generated two QKI5 constructs carrying either a V157E mutation (pCMV6-QKI5V157E) or a deletion of the KH RNA-binding domain (pCMV6-QKI5ΔKH) to assess their effects on the output of pri-124-1 and miR-124 from a plasmid (pCMV6-124_WT) in which human miR-124-1 is transcribed from the cytomegalovirus (CMV) promoter (Figure 3A). q-PCR results showed that the mutant QKI5V157E or QKI5ΔKH abolished the effects caused by wild-type QKI5 (Figure 3B, Supplementary information, Figure S3A and S3B). Moreover, when the QRE sequence on pri-124-1 was mutated (pCMV6-124_MUT), even wild-type QKI5 failed to regulate the level of the CMV-transcribed mutant pri-124-1 and miR-124 (Figure 3B). Thus, the pri-124-1 processing was regulated by QKI5 binding.
QKI5 binds to the pri-124-1 transcript in vitro and in vivo. (A) A schematic representation of co-transfection with wild-type (124_WT) or mutant (124_MUT) pri-124-1 constructs and either wild-type (QKI5) or mutant (QKIΔKH and QKI5V157E) QKI5 constructs. The nucleotide positions of the QRE and stem loop are indicated in the cloned pri-124-1 sequence used in this study. (B) q-PCR shows the changes in the levels of pri-124-1 (red bar) and miR-124 (green bar) upon transfection of the indicated combinations into 293T cells as shown in A. (C) A schematic representation of the primers used in RNA-IP-PCR analysis. (D) RNA-IP-PCR assays for pri-124-1 performed on anti-QKI5 immunoprecipitates (QKI5) from K562 and HEL cell lysates. An unrelated IgG served as the negative control (IgG). RT: no reverse-transcribed PCR. QKI5 RNA-IP assays for the irrelevant pri-23a are shown in the right panel. (E) q-PCR analysis of pri-124-1 and pri-23a associated with QKI5 evaluated by RNA-IP assays in K562 cells. The RNA-IP-q-PCR results are shown as fold enrichment compared with input. (F) A schematic representation of the DNA oligonucleotides used in the RNase H protection assays. The antisense DNA oligonucleotides A-C span different regions of 124-QRE RNAs, whereas Ctr is an unrelated control oligonucleotide. (G, H) RNase H protection assays. RNase H cleavage results of reaction with either 124-QRE RNAs or QKI5-protected 124-QRE RNAs targeted by antisense DNA oligonucleotides A-C (lanes 2-4 and 7-9) or by an unrelated control oligonucleotide (Ctr, lanes 5 and 10; G). Additional control reactions were performed in the absence of oligonucleotide (input, lanes 1 and 6). Arrow indicates a reduced cutting site (G). The quantitative data were shown in H. The relative protection ratio was defined by the percentage of undigested 124-QRE in reactions with QKI5 protein to that without QKI5, and the relative digestion ratio was defined by the percentage of digested segments in reactions with QKI5 protein to that without QKI5. The data were quantified using ImageJ software. (I) In vitro association of QKI5 with the pri-124-1 QRE as identified by an RNA-EMSA assay in which 5′-biotin-labeled miR-124-QRE probes were incubated with different concentration of purified Flag-QKI5 (the left panel). A reported 5′-biotin-labeled p27-QRE probe was used as the positive control (the right panel). A mutant probe was used as the control, and the unlabeled miR-124-QRE probe was used in the competitive assays. (J) A schematic representation of the RNA pull-down assays using MS2-tagged pri-124-1 affinity purification. (K) Immunoblot of endogenous QKI5 in RNA pull-down assays from 293T cells transfected with either MS2-tagged wild-type pri-124-1 (124_WT-MS2) or MS2-tagged QRE mutant pri-124-1 (124_MUT-MS2) constructs. An unrelated protein GAPDH was used as the control. (L) The competitive association of non-MS2-tagged pri-124-1 with endogenous QKI5 in RNA pull-down assays performed on 293T cells co-transfected with 124_WT-MS2 and increased dosages of non-tagged 124_WT. Error bars reflect SEM from three biological replicates if not stated otherwise. Significance was determined by t-test with *P< 0.05; **P< 0.01; ***P< 0.001.
QKI5 is capable of binding to pri-124-1 in vitro and in vivo
To further confirm the physical interactions between QKI5 and pri-124-1, RNA-immunoprecipitation (RIP)-PCR with three sets of primers (Figure 3C) was conducted and showed that QKI5 could bind endogenous pri-124-1 but not pri-23a∼27a∼24-2 in K562 and HEL cells (Figure 3D and 3E). To determine whether the QRE motif in pri-124-1 is a bona fide QKI-binding site, RNase H protection assay was performed by incubating 124-QRE RNAs (174 nt) harboring the predicted QRE with different DNA oligonucleotides A-C (Figure 3F). All three probes (A-C) were able to target RNase H-mediated cleavage to none protein-incubated RNAs, whereas in control reactions done in the presence of an unrelated oligonucleotide (Ctr), 124-QRE RNA remained intact (Figure 3G). In contrast, the digestion efficiency of QKI5-preincubated RNA was not affected by oligonucleotides B-C, but was affected by oligonucleotide A (Figure 3G and 3H). Thus, oligonucleotide A spanning the predicted QRE motif protected the RNA component from probe-targeted RNase H digestion to some extent, confirming QKI5's binding on this site (Figure 3H).
To learn more about the interaction between QKI5 and pri-124-1, RNA electrophoretic mobility shift assay (EMSA) was further performed using purified recombinant QKI5 protein. P27 mRNA, which had been reported to be bound and stabilized by QKI, was used as a positive control24. The slow-migrating QKI5/miR-124-QRE (a 29 nt RNA probe harboring the QRE motif of pri-124-1) complexes formed in a dose-dependent manner with the addition of QKI5 protein (Figure 3I). In addition, a QRE-mutant probe could not efficiently bind QKI5, and an unlabeled probe sufficiently out-competed the slow-migrating complexes, confirming the specific interaction between QKI5 and pri-124-1 in vitro (Figure 3I).
Subsequently, we performed RNA pull-down analysis by generating a new MS2-tagged wild-type pri-124-1 construct (124_WT-MS2, in which the MS2 hairpins could bind to the MS2 coat protein) and another QRE-mutant pri-124-1 construct (124_MUT-MS2; Figure 3J). 293T cells expressing MS2-tagged pri-124-1 exhibited an ∼200-fold higher level of pri-124-1 than the empty constructs (Supplementary information, Figure S3C). The addition of MS2 (MS2 coat protein)-MBP (maltose-binding protein) fusion protein to the lysates with amylose resin recovered up to 30% of pri-124-1 RNAs (Supplementary information, Figure S3D). More importantly, QKI5 could be co-purified with the wild-type MS2-tagged pri-124-1 but not the QRE-mutant pri-124-1 (Figure 3K); the irrelevant protein GAPDH was used as a negative control (Figure 3K). The interaction specificity was further examined by a competitive RNA pull-down experiment. QKI5 binding to pri-124-1 was efficiently and competitively inhibited by increasing the amounts of non-MS2-tagged pri-124-1 (Figure 3L and Supplementary information, Figure S3D). Taken together, these data suggest that pri-124-1 RNA represents a bona fide target for QKI5.
QKI5 is associated with Microprocessor (DGCR8/Drosha) complex
Microprocessor initiates miRNA processing and defines the abundance of certain pri-miRNAs and miRNAs. The above results prompted us to examine whether QKI5 regulates pri-124-1 processing through interacting with this complex (DGCR8/Drosha). To test this possibility, K562 and HEL extracts were incubated with either Flag-QKI5- or Flag-RFP-coated beads, and the results showed that DGCR8 and Drosha specifically bound to QKI5 but not the RFP protein (Figure 4A). To further examine whether QKI5 is associated with Microprocessor in vivo, co-IP experiment was performed and showed that the endogenous QKI5 could interact with DGCR8 and Drosha in an RNA-independent manner (Supplementary information, Figure S3E). The subsequent protein fractionation experiments by fast protein liquid chromatography (FPLC) further indicated that native QKI5 from K562 cell nuclear extracts (NEs) was detected in chromatographic fractions from a Superose 6 column and had a relatively symmetrical peak of ∼440 kDa (Figure 4B). Interestingly, the elution pattern of QKI5 overlapped with that of Drosha and DGCR86, supporting the idea that endogenous QKI5 may also be associated with Microprocessor. The subsequent immunoprecipitation (IP) assay confirmed this in vivo interaction in K562 and HEL cells (Figure 4C). In addition, double labeling of K562, HEL and 293T cells using QKI5 mouse monoclonal and Drosha or DGCR8 rabbit polyclonal antibodies revealed their co-localization in the nucleus (Figure 4D).
QKI5 interacts with Microprocessor complex. (A) Identification of the interaction of QKI5 with Drosha and DGCR8 by affinity purification. 293T cells were transfected with a construct encoding Flag-QKI5 or Flag-RFP, and then Flag-tagged bait proteins were recovered from cell lysates on Flag-agarose beads. Whole-cell lysates from K562 or HEL cells were subjected to affinity purification with the indicated bait. Bound proteins were eluted from the beads and immunoblotted with the indicated antibodies. (B) Co-fractionation of QKI5 and Microprocessor by FPLC. Nuclear extracts from K562 cells were fractionated on a Superose 6 gel filtration column. Chromatographic elution profiles and immunoblot analysis of the chromatographic fractions are shown. The positions of the molecular mass size markers are indicated by arrowheads, and an equal volume from each fraction was analyzed. (C) QKI5 interacts with the DGCR8/Drosha complex in vivo. Whole K562 or HEL cell lysates were immunoprecipitated with anti-QKI5. Immunocomplexes were then immunoblotted using antibodies against the indicated proteins. An unrelated protein GAPDH was used as the control. (D) Immunofluorescence analysis of the colocalization of QKI5 with Drosha or DGCR8 in K562, HEL and 293T cells. The cells were immunostained with anti-QKI5 and anti-Drosha or anti-DGCR8 and labeled with fluorescent secondary antibodies. Red, QKI5; green, Drosha or DGCR8; blue, nucleus. Scale bar, 30 μm. (E) QKI5 specifically interacts with DGCR8 in vitro. Immobilized GST or GST-QKI5 was incubated with 293T cell lysates expressing Flag-Drosha or Flag-DGCR8. Bound proteins were eluted from the beads and immunoblotted with anti-Flag. The amounts of GST and GST-QKI5 were determined by immunoblotting with anti-GST. (F) Mapping of the DGCR8 and QKI5 domains involved in their interaction. Left panel: the domain structures of DGCR8 and its deletion mutants are shown. Immobilized GST-QKI5 was incubated with 293T cell lysates expressing full-length DGCR8 or one of the Flag-DGCR8 deletion mutants. Right panel: the domain structures of QKI5 and its deletion mutants are shown. Immobilized GST-QKI5 or its deletion mutants were incubated with 293T cell lysates expressing Flag-DGCR8.
To characterize the interactions of these proteins more precisely, GST pull-down assay was performed by incubating lysates of 293T cells expressing exogenous Flag-Drosha or Flag-DGCR8 with immobilized GST or GST-QKI5 proteins. Immunoblots of pull-down samples indicated that QKI5 directly associated with DGCR8, but not Drosha (Figure 4E). Furthermore, to determine the protein interaction interface of QKI5 and DGCR8, GST pull-down assays were performed with the GST-QKI5 protein and Flag-tagged full-length (1-773 aa), ΔC263 (1-510 aa), ΔC473 (1-300 aa), ΔN510 (511-773 aa) and ΔN300 (301-773 aa) DGCR8 constructs expressed in 293T cells. The results showed that the double DRSM domain of DGCR8 is responsible for the interaction of DGCR8 with QKI5 (Figure 4F). In addition, we mapped the DGCR8 interaction interface of QKI5 using GST fusions of the full length (1-341 aa), N-terminal fragment (1-180 aa) and C-terminal fragment (181-341 aa) of QKI5. Our results suggest that the KH domain of QKI5 is responsible for their interaction (Figure 4F).
QKI5 increases Microprocessor recruitment to pri-124-1
To test whether the QKI5/DGCR8/Drosha complex assembled specifically on pri-124-1, RNA pull-down analysis was performed with either MS2-tagged wild-type pri-124-1 (124_WT-MS2) or QRE-mutant pri-124-1 (124-MUT-MS2; Figure 5A). The associations of QKI5, DGCR8 and Drosha with pri-124-1 were dramatically enriched when the QRE motif was present (Figure 5A). Similar to QKI5, the co-precipitation of DGCR8 and Drosha was competitively reduced by adding non-MS2-tagged pri-124-1 (Figure 5B). Furthermore, ectopic expression of QKI5 increased the association of DGCR8 and Drosha with pri-124-1, whereas reduced expression of QKI5 decreased their recruitment to pri-124-1 RNAs (Figure 5C). As the endogenous levels of DGCR8 and Drosha proteins were relatively constant upon QKI5 overexpression or knockdown (Figure 5C), the mechanism by which QKI5 activated pri-124-1 processing might primarily rely on its regulation of the recruitment efficiency of Microprocessor. These results indicate that QKI5 recognizes the QRE on pri-124-1 and increases Microprocessor recruitment by interacting with DGCR8.
Association of QKI5 with pri-124-1 promotes its processing by Microprocessor. (A) Immunoblot of endogenous QKI5, DGCR8 and Drosha in RNA pull-down assays from 293T cells transfected with either MS2-tagged 124_WT (124_WT-MS2) or MS2-tagged 124_MUT (124_MUT-MS2). The unrelated protein GAPDH was used as a control. The same sample as shown in Figure 3K was used in this analysis. (B) Immunoblot of endogenous QKI5, DGCR8 and Drosha in competitive RNA pull-down assays, as shown in Figure 3J. The same sample as shown in Figure 3L was used in this analysis. (C) Left WB panels: immunoblot of endogenous levels of QKI5 (or Flag-QKI5), DGCR8 and Drosha in 293T cells co-transfected with 124_WT-MS2 and either pCMV6-QKI5 or pCMV6, and si-QKI5 or si-control. Right RNA pull-down panels: immunoblot of endogenous QKI5, DGCR8 and Drosha associated with MS2-tagged 124_WT in RNA pull-down assays. (D) In vitro processing assay using [α-32P] UTP-labeled wild-type pri-124-1 (124_WT) or QRE mutant pri-124-1 (124_MUT) as the substrates. Pri-miRNA transcripts are incubated with QKI5-depleted HeLa NEs and various amounts of purified Flag-QKI5 as described in Materials and Methods. The sequences of wild-type and mutant QRE motifs are shown at the bottom of the figure, and altered sequences are indicated by an underline. (E) The quantitative data were derived from three independent in vitro processing experiments. The calculation formula was shown in Supplementary information, Figure S4B. (F) RNA-RNA interaction detected by band shift experiments. Upper panel: visualization of the predicted duplexes formed by 4-26 nt (included in the R1 region, 1-48 nt) and 552-572 nt (included in the R2 region, 542-580 nt) of pri-124-1 (for binding details, see Supplementary information, Figure S6C). Lower panel: band shift experiments with biotin-labeled wild-type R2 (R2_WT) or mutant R2 (R2_MUT) and increasing volumes of unlabeled wild-type R1 (R1_WT) or mutant R1 (R1_MUT, a complementary mutation to R2_MUT). The quantitative data were derived from 2-3 independent experiments. (G) A schematic representation of pri-124-1 variants that have wild-type or mutant R1 or/and R2 sequence. (H) The association of QKI5, DGCR8 and Drosha with different MS2-tagged pri-124-1 variants as described in G. (I) In vitro processing assay using [α-32P] UTP-labeled pri-124-1 variants as described in G. (J) The quantitative results of in vitro processing assay in I. The calculation formula was shown in Supplementary information, Figure S4B. Error bars reflect SEM from three biological replicates if not stated otherwise. Significance was determined by t-test with *P< 0.05.
QKI5 promotes pri-124-1 processing in vitro and in vivo
We next asked whether QKI5 could modulate Microprocessor function on pri-124-1 processing in vitro. To avoid influence of QKI5 protein in HeLa cell NEs, we first performed QKI5 depletion by a specific QKI5 antibody. Then the transcribed wild-type (124_WT) or QRE-mutant (124_MUT) pri-124-1 substrates were labeled with 32P and subjected to processing by QKI5-depleted NEs. The addition of increased amounts of QKI5 protein increased the processing efficiency of 124_WT, as indicated by a higher amount of pre-124-1 products (Figure 5D and 5E). In contrast, QKI5 failed to affect the processing of either 124_MUT or a control pri-23a∼27a∼24-2 to corresponding precursor miRNAs (Figure 5D and 5E, Supplementary information, Figure S4A). To further verify the above findings in vivo, GFP-labeled wild-type (GFP-124_WT) or QRE-mutated (GFP-124_MUT) pri-124-1 constructs (Supplementary information, Figure S4C) were co-transfected with pCMV6-QKI5 into 293T cells, respectively. The introduction of QKI5 significantly attenuated the fluorescence of GFP-124_WT, whereas the fluorescence of GFP-124_MUT was unaffected by ectopic QKI5 expression (Supplementary information, Figure S4C). These data suggest that QKI5 could increase the processing of pri-124-1 carrying a functional QRE motif.
QKI5 promotes pri-124-1 processing at a distance
As the identified QRE is localized ∼300 nt upstream of the pri-124-1 stem loops, we aimed to determine how a distal RNA motif facilitates its processing. In cells, despite being single-stranded, RNA molecules often fold back on themselves to form base-paired structures between short stretches of complementary sequences; thus, sites separated by many base pairs may not actually be as far apart as previously thought. In addition, the direct RNA-RNA interaction may actively bring two RNA segments into close proximity. To computationally predict the possible interactions between the QRE and pri-124-1 stem loop regions, we used the IntaRNA webserver from http://rna.informatik.uni-freiburg.de:8080/v1/IntaRNA.jsp25. A highly predicted RNA-RNA interaction region (R1:R2) was localized adjacent to the QRE motif and near downstream of the pri-124-1 stem loops (Figure 5F and Supplementary information, Figure S4D). To test this possibility, we performed band shift experiments with four different in vitro synthesized RNAs: wild-type R1 and R2 (R1_WT and R2_WT), as well as mutant R1 and R2 in which the predicted base-paired stem is disrupted (R1_MUT and R2_MUT) (Figure 5F and Supplementary information, Table S4). In the first round, biotin-labeled R2_WT was incubated with increasing concentrations of R1_WT or R1_MUT, and band shifts were observed with R1_WT but not with R1_MUT (Figure 5F). Similarly, R2_MUT was not able to interact with R1_WT (Figure 5F). Nevertheless, the compensatory mutation (R1_MUT), which was able to reform a base-paired stem with different primary sequences, still retarded the R2_MUT RNAs (Figure 5F).
To further determine whether this RNA-RNA interaction was responsible for QKI5 regulation, constructs expressing one or two of the variants (124-R1_MUT, 124-R2_MUT or 124-R1+2_MUT), as well as 124_WT (Figure 5G) were used in MS2-tagged RNA pull-down (Figure 5H) and in vitro processing assays (Figure 5I and 5J). First, the associations of QKI5, DGCR8 and Drosha with the four pri-124-1 transcripts were not affected no matter whether the R1:R2 interaction existed or not (Figure 5H). However, disruption of this interaction (mutation in R1 or R2 alone: 124-R1_MUT or 124-R2_MUT, respectively) was sufficient to abrogate QKI5 function in in vitro processing assays (Figure 5I and 5J). Nevertheless, this regulation did not reflect a requirement for the primary RNA sequence because the compensatory mutation (124-R1+R2_MUT) restored full QKI5 activity in pri-124-1 processing assay (Figure 5I and 5J). Collectively, these results suggest that both Microprocessor recruitment and R1:R2 interactions were required for QKI5 function. Through the spatial RNA-RNA interaction, the QKI5-recruited Microprocessor reaches the pri-124-1 stem loops and facilitates its processing.
Given the significance of QRE for QKI5's ability to regulate pri-124-1 cleavage, we hypothesized that other pri-miRNAs harboring a QRE might also be reasonable targets of QKI5. To investigate this possibility, we introduced a QRE either upstream or downstream of pri-23a∼27a∼24-2 (Supplementary information, Figure S5A). As expected, the mature miRNAs expressed from both 23a_QREup and 23a_QREdown were induced by ∼3-fold upon ectopic QKI5 expression, whereas pri-miRNA was reduced by ∼5-fold, indicating that processing of the QRE-containing pri-23a∼27a∼24-2 was promoted (Supplementary information, Figure S5B). In addition, RNA pull-down analysis using MS2-tagged wild-type or QRE-containing pri-23a∼27a∼24-2 was performed to validate whether the regulation is attributed to QKI5-mediated recruitment of the Microprocessor. Immunoblot of RNA precipitates showed that DGCR8 and Drosha were indeed recruited more strongly to QRE-containing pri-23a∼27a∼24-2, regardless of whether the QRE was inserted upstream or downstream of miRNA stem loops (Supplementary information, Figure S5C and S5D). Thus, the action of the QRE-containing pri-23a∼27a∼24-2 appeared to mimic the effect of QKI5 on pri-124-1 in vitro.
Attenuation of miR-124 and QKI5 is required for erythroid differentiation
QKI5 was initially identified as a candidate for the classical neurologic mutation qkv, which is related to glial cell differentiation26,27 as well as various developmental processes; however, little is known regarding its role in erythroid programming. In our experiments, the level of QKI5 continuously decreased during erythroid differentiation of HPCs (Figure 2B), indicating its involvement in this process. In addition, the expression of QKI5 paralleled that of miR-124 during this process, further indicating a physiological connection between these regulators. To examine the relevance of these regulators (QKI5 and miR-124) in erythroid differentiation, HPCs were first transduced with lentivirus expressing GFP (lenti-GFP), miR-124 (lenti-124) or miR-124 inhibitors (lenti-Zip-124), and then induced by EPO to erythroid maturation. Given the decreased processing efficiency of pri-124-1 during erythroid differentiation, we also generated an alternative lentivirus construct by placing the ∼80-nt hairpin pre-124-1 under the control of RNA polymerase III (pSIH-124). As shown in Supplementary information, Figure S6A and S6C, the level of miR-124 that matured from pSIH-124 (∼3-fold increase) was less than that processed from lenti-124. Therefore, we still chose the lenti-124 construct for further investigation.
After HPCs were cultured for 7 days in EPO-supplemented medium, the HPCs could be roughly classified into three differentiation stages according to the abundance of erythroid markers CD235a and CD71: undifferentiated erythroid progenitors (CD71−/CD235a−), moderately differentiated erythrocytes (CD71high/CD235a+) and more mature erythrocytes (CD71low/CD235a+). Of note, ectopic miR-124 reduced the kinetics of HPC erythroid maturation, as shown by decreased numbers of differentiated CD71high/CD235a+ cells but increased numbers of CD71−/CD235a− progenitors on days 7 and 11 of erythroid culture (Figure 6A and Supplementary information, Figure S6D). In fact, the miR-124-transduced HPCs had higher percentage of immature basophilic erythroblasts on day 7 of culture (Figure 6B and 6C), and remained as immature erythroblasts (pro-erythroblasts and basophilic erythroblasts) after 11 days (Figure 6C and Supplementary information, Figure S6D). Accordingly, inhibition of miR-124 (Supplementary information, Figure S6A) augmented HPC erythroid differentiation (Figure 6D-6F, Supplementary information, Figure S6E), suggesting a negative role for miR-124 in erythropoiesis.
miR-124 and QKI5 are negative regulators of erythroid differentiation. (A) Monitoring of the viable HPC population (left panel) and the CD235a-/CD71-stained fraction (medium panel) of lenti-GFP- or lenti-124-transduced HPCs on day 7 of E culture. The morphology (May-Grunwald Giemsa staining) of HPCs is shown in the right panel. A 400× magnification of a representative field is shown. Scale bar, 50 μm. (B) Quantification of the percentage of cells gated as CD71−/CD235a−, CD71high/CD235a+ and CD71low/CD235a+ fractions in HPCs transduced with lenti-GFP or lenti-124 on days 7 and 11 of E culture. (C) Quantification of the percentages of basophilic (Bas), polychromatophilic (Pol), orthochromatic (Ort) erythroblasts and erythrocytes (Ery) that were determined by Giemsa staining of cytospin preparations of lenti-GFP- or lenti-124-transduced HPCs on days 7 and 11 of E culture. (D) Monitoring of the viable HPC population (left panel) and the CD235a-/CD71-stained fraction (medium panel) of lenti-GFP- or lenti-Zip-124-transduced HPCs on day 7 of E culture. The morphology (May-Grunwald Giemsa staining) of HPCs is shown in the right panel. A 400× magnification of a representative field is shown. Scale bar, 50 μm. (E) Quantification of the percentage of cells gated as CD71−/CD235a−, CD71high/CD235a+ and CD71low/CD235a+ fractions in HPCs transduced with lenti-GFP or lenti-Zip-124 on days 7 and 11 of E culture. (F) Quantification of the percentages of basophilic (Bas), polychromatophilic (Pol), orthochromatic (Ort) erythroblasts and erythrocytes (Ery) that were determined by Giemsa staining of cytospin preparations of lenti-GFP- or lenti-Zip-124-transduced HPCs on days 7 and 11 of E culture. (G) Monitoring of the viable HPC population (left panel) and the CD235a-/CD71-stained fraction (medium panel) of lenti-GFP- or lenti-si_QKI5-transduced HPCs on day 7 of E culture. The morphology (May-Grunwald Giemsa staining) of HPCs is shown in the right panel. A 400× magnification of a representative field is shown. Scale bar, 50 μm. (H) Quantification of the percentage of cells gated as CD71−/CD235a−, CD71high/CD235a+ and CD71low/CD235a+ fractions in HPCs transduced with lenti-GFP or lenti-si_QKI5 on days 7 and 11 of E culture. (I) Quantification of the percentages of basophilic (Bas), polychromatophilic (Pol), orthochromatic (Ort) erythroblasts and erythrocytes (Ery) that were determined by Giemsa staining of cytospin preparations of lenti-GFP- or lenti-si_QKI5-transduced HPCs on days 7 and 11 of E culture. (J) Monitoring of the viable HPC population (left panel) and the CD235a-/CD71-stained fraction (medium panel) of lenti-GFP- or lenti-QKI5-transduced HPCs on day 7 of E culture. The morphology (May-Grunwald Giemsa staining) of HPCs is shown in the right panel. A 400× magnification of a representative field is shown. Scale bar, 50 μm. (K) Quantification of the percentage of cells gated as CD71−/CD235a−, CD71high/CD235a+ and CD71low/CD235a+ fractions in HPCs transduced with lenti-GFP or lenti-QKI5 on days 7 and 11 of E culture. (L) Quantification of the percentages of basophilic (Bas), polychromatophilic (Pol), orthochromatic (Ort) erythroblasts and erythrocytes (Ery) that were determined by Giemsa staining of cytospin preparations of lenti-GFP- or lenti-QKI5-transduced HPCs on days 7 and 11 of E culture. Error bars reflect SEM from three biological replicates if not stated otherwise. Significance was determined by t-test with *P< 0.05.
In addition, in the gain- or loss-of-function analysis of QKI5 (Supplementary information, Figure S6B), the percentage of differentiated CD71high/CD235a+ cells was decreased by ∼10% on day 7 and by ∼20% on day 11 in QKI5-overexpressing HPCs (Figure 6J-6L, Supplementary information, Figure S6F), whereas the erythroid maturation was promoted in QKI5-knockdown HPCs (Figure 6G-6I, Supplementary information, Figure S6G), indicating an inhibitory role similar to that of miR-124 in erythropoiesis.
Furthermore, re-introduction of miR-124 could restore the erythroid maturation of HPCs driven by QKI5 knockdown (Supplementary information, Figure S7A), whereas suppression of miR-124 compensated for the impaired erythroid differentiation caused by QKI5 overexpression (Supplementary information, Figure S7B). These results suggest that the differentiation-inhibiting effect of miR-124 is closely connected to QKI5 regulation in HPCs.
miR-124 inhibits erythroid differentiation through targeting c-MYB and TAL1
In general, QKI5 regulates erythropoiesis, at least in part, via its ability to activate pri-124-1 processing, however, the downstream effectors of this QKI5-miR-124 pathway remain unknown. To investigate the downstream targets of miR-124, the miRanda, PicTar and TargetScan databases were utilized. Among the predicted targets, c-MYB and TAL1 (Figure 7A) are well-known positive regulators of erythroid differentiation28,29. To examine whether the antagonistic effect of miR-124 on erythropoiesis is dependent on the miRNA:target interactions, we first cloned the wild-type (wild-type reporter) or miR-124-binding site-mutant (mutant reporter) c-MYB and TAL1 3′ untranslated regions (UTRs; Figure 7A) into a pMIR-reporter plasmid and co-transfected these constructs into 293T cells with miR-124 mimic or a scrambled control, respectively. Reporter assays revealed that miR-124 significantly reduced the luciferase activities of wild-type c-MYB and TAL1 reporters compared with the scrambled control (Figure 7B). In contrast, the luciferase activities of the mutant reporters were not repressed by miR-124, indicating that the repression was dependent on miRNA binding (Figure 7B). Furthermore, c-MYB and TAL1 were downregulated by ectopic miR-124 in HPCs, and their upregulation was observed after QKI5 repression, which reduced the level of miR-124 (Figure 7C). Similarly, the endogenous levels of c-MYB and TAL1 were reduced in miR-124 mimic-transfected K562 cells but not in control-transfected K562 cells (Supplementary information, Figure S8A), and downregulating QKI5 ultimately increased the intracellular levels of c-MYB and TAL1 (Supplementary information, Figure S8A). Thus, the differentiation-inhibitory activity of QKI5 or miR-124 was likely mediated by their indirect or direct repression of c-MYB and TAL1 expression during erythropoiesis.
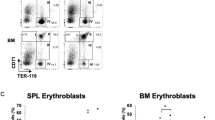
The QKI5-miR-124-targets axis in erythroid cells. (A) A computer prediction of the conserved and mutated binding sites within the 3′ UTR of human c-MYB and TAL1 mRNAs for miR-124. (B) Relative luciferase activity of the indicated reporter constructs. (C) Immunoblot of endogenous c-MYB and TAL1 levels in HPCs transduced with lenti-124 or lenti-si_QKI5. (D) Monitoring of CD235a-/CD71-stained fraction in bone marrow (BM) from mice transplanted with lenti-GFP-transduced HPCs (GFP mice), lenti-QKI5-transduced HPCs (QKI5 mice) or lenti-124-transduced HPCs (miR-124 mice). (E) Quantification of the percentage of cells gated as CD71−/CD235a−, CD71high/CD235a+ and CD71low/CD235a+ fractions of human HPC-derived cells in BM and spleen (SP) from GFP mice, QKI5 mice and miR-124 mice. (F) q-PCR of pri-124-1 and miR-124 levels in the BM and SP of transplanted mice. (G) Immunoblot of QKI5, c-MYB and TAL1 protein levels in the BM of transplanted mice. Error bars reflect SEM from three biological replicates if not stated otherwise. Significance was determined by t-test with *P< 0.05; **P< 0.01.
QKI5 and miR-124 negatively regulate erythropoiesis in xenograft mice
To further determine the functional relevance of QKI5 and miR-124 in vivo, NOD/SCID mice (n = 3 per group) were intravenously injected with human HPCs transduced with either lenti-GFP (GFP mice), lenti-QKI5 (QKI5 mice) or lenti-124 (miR-124 mice) to establish engraftment models (Supplementary information, Figure S7B). The mice were assessed 4 weeks later by flow cytometric quantification of a human specific myeloid marker, CD33, in two hematopoietic organs, bone marrow (BM) and spleen (SP) (Supplementary information, Figure S8C and S8D). An ∼3-fold increase in miR-124 was observed in BM and SP from miR-124 mice than the control GFP mice (Supplementary information, Figure S7E), whereas the QKI5 protein level was increased by 2-3-fold in BM from QKI5 mice (Supplementary information, Figure S8F). The resulting human erythroid differentiation was measured by staining for CD235a and CD71 in CD33+ human cells. QKI5 and miR-124 mice showed decreased numbers of differentiated CD71high/CD235a+ cells in both BM and SP cell components (Figure 7D and 7E, Supplementary information, Figure S8G) compared with the GFP mice. In addition, histologic analysis of the mouse SPs revealed a greater number of erythroid cell clusters in the QKI5 and miR-124 mice, indicating impaired erythroid maturation (Supplementary information, Figure S8H). Thus, these results confirm the negative roles of QKI5 and miR-124 in erythropoiesis in vivo.
Furthermore, in both the BM and SP from QKI5 mice, the level of human pri-124-1 was downregulated by ∼3-fold, whereas miR-124 was upregulated by ∼2-fold (Figure 7F). In this case, although mmu-miR-124 could not be distinguished from the total miR-124, the influence of QKI5 on mouse pri-124 processing (a conserved QRE is located ∼300 nt upstream of mouse pri-124-2 stem loops) would most likely amplify this result.
In addition, enforced miR-124 expression in BM markedly reduced c-MYB and TAL1 levels (Figure 7G), and QKI5 overexpression had the same effect (Figure 7G). Altogether, these in vitro and in vivo results indicate that reduced QKI5 expression during erythroid differentiation contributes to the post-transcriptional attenuation of the erythropoiesis antagonist miR-124; this subsequently releases the expression of two critical erythropoiesis modulators, c-MYB and TAL1.
Global regulation of miRNA production by QKI5
The above results demonstrate that downregulation of QKI5 contributes to the post-transcriptional attenuation of differentiation-inhibitory miR-124, an event required for normal erythroid development. We then asked whether other miRNAs, particularly those miRNAs antagonistic to erythropoiesis, could also be modulated by QKI5. To investigate this hypothesis, global miRNA expression was assessed upon QKI5 overexpression. Total RNA samples were prepared from GFP+-selected K562 cells (Supplementary information, Figure S9A) with ectopic QKI5 expression (QKI5) or GFP transduction (Con; Supplementary information, Figure S9B). Small RNAs in the miRNA size range were examined by deep sequencing. The size distribution peaked at ∼22 nt (Supplementary information, Figure S9C), and the number of miRNA reads (Figure 8A) did not differ between the control and QKI5-overexpressed K562 cells. To identify differentiation-inhibitory miRNAs that were regulated by QKI5 similarly to miR-124, we compared upregulated miRNAs from QKI5 overexpression profiles to our previous miRNA array data of K562 erythroid differentiation and discovered 44 candidates (Figure 8B and Supplementary information, Table S5). To confirm miRNA expression independently and more quantitatively, we selected 14 candidates (Figure 8C) with comparable enrichment in K562 cells and determined their expression by q-PCR. Except for miR-501-5p, all of the miRNAs showed continuously decreased expression during erythroid differentiation (Figure 8C). However, we observed the accumulation of pri-34a, -331, -454, -574, -135a and -130b after hemin treatment (Figure 8C), suggesting the existence of post-transcriptional regulation of these pri-miRNAs. Furthermore, q-PCR analysis of QKI5-overexpressing or QKI5-knockdown K562 cells indicated that miR-34a-5p, -454-3p, -574-3p, -130b-3p and -135a-5p (hereinafter called miR-34a, -454, -574, -130b and -135a) are candidate targets of QKI5 (Figure 8C and Supplementary information, Table S6).
QKI5 regulates multiple miRNA targets in erythroid cells. (A) Comparison of miRNA abundance in small RNA-seq data from QKI5 versus control. (B) Heatmap representation of relative miRNA expression obtained from small RNA-seq data (left panel) and microarray data (right panel, the miRNA array was performed in K562 cells undergoing erythroid differentiation by hemin treatment, see also Supplementary information, Table S5). (C) Heatmap representation of q-PCR results of pri-miRNA and miRNA transcripts corresponding to individual miRNAs either in K562 cells of E culture (left panel), K562 cells with QKI5 overexpression (medium panel) or K562 cells with endogenous QKI5 knockdown (right panel). (D) The left panel: representation of the predicted QKI5 binding sites (QRE, red bar) on possible pri-miRNA transcripts spanning ± 2 kb fragments from miRNA stem loops (round black). The medium panel: RNA-IP-PCR assays for pri-miRNAs performed on anti-QKI5 immunoprecipitates (QKI5) from K562 cell lysates. RNA-IP with unrelated IgG serves as the control (IgG). RT: no reverse-transcribed PCR reaction. The right panel: the genomic location of each miRNA. (E) A schematic representation of target genes overlapping between miR-124, -130, -135 and -454. (F) Hypergeometric test suggesting a significant overlap between miRNA target genes and “erythroid genes” in dataset GSE43023. (G) A schematic representation of how QKI5 acts at a distance to regulate Microprocessor-mediated pri-124-1 processing during erythropoiesis.
Furthermore, to verify whether QKI5-mediated regulation is due to its recognition of pri-miRNAs, we predicted potential QREs located within 2 kb of pri-miRNA stem loops in the RBPDB database and performed RNA-protein complex immunoprecipitation (RNA-IP)-PCR analysis to determine their interactions (Figure 8D). Although the full-length transcripts of individual pri-miRNAs need to be further identified, RNA fragments within 2 kb of the miR-34a, -454, -574, -135a and -130b stem loops were found to be associated with endogenous QKI5 in K562 cells (Figure 8D). Taken together, our findings suggest that QKI5 may bind to multiple pri-miRNAs and modulate their processing, which likely accounts for the post-transcriptional regulation of these miRNAs in differentiating erythroid cells.
QKI5-responsive miRNAs have a signature of erythroid-regulatory activity
To determine whether these identified QKI5-responsive miRNAs are involved in erythroid differentiation, we used K562 and HEL cells (robust in vitro models) to assess their function by detecting the surface markers CD235a/CD71 (Supplementary information, Figure S9D), staining for hemoglobin with benzidine (DAB; Supplementary information, Figure S9E) and quantifying γ-globin mRNAs (Supplementary information, Figure S9F). As a control, the well-studied erythropoiesis antagonist miR-223-3p (hereinafter called miR-223) was included. The miR-223-, -454-, -130b- or -135a-expressing K562 cells were blocked from differentiation as indicated by the decrease in CD71+/CD235a+ cells after 48-h hemin treatment (Supplementary information, Figure S9D). In addition, these cells exhibited a reduced percentage of DAB-positive cells (Supplementary information, Figure S9E) and lower γ-globin mRNA levels (Supplementary information, Figure S9F). The functional analysis of these miRNAs revealed a similar result in HEL cells (Supplementary information, Figure S9D-S9F). Therefore, these data indicate that the QKI5-associated post-transcriptional attenuation of miR-454, 135a and -130b, as well as miR-124, may be a common paradigm required for normal erythroid development.
To gain further insights into the biological functions of these miRNAs, we performed computational analyses to identify overlaps or synergy between miRNA target profiles and to predict their changes during erythropoiesis. First, the highly reliable target genes of miR-124, -454, -135a and -130b were obtained from miRTarBase and starBase, respectively. Notably, target genes with the annotation “weak interactions” in miRTarBase were excluded, and only those supported by at least five experiments and validated by at least four prediction tools in starBase were adopted (Figure 8E and Supplementary information, Table S7). Gene Ontology analysis showed that miR-124, -454, -135a or -130b synergistically regulate cohorts of genes that participate in similar processes, including hematopoiesis and hematopoietic or lymphoid organ development (Supplementary information, Table S7). More importantly, common targets of these miRNAs were also enriched for erythropoiesis (Figure 8E and Supplementary information, Table S7).
Next, to predict the erythroid-regulatory activity of these miRNA target genes, four erythropoiesis-associated RNA-Seq or array datasets (GSE43023, GSE53635, GSE61566 and GSE53983) were downloaded from the GEO database. A group of “erythroid genes” was defined by both high expression and upregulation along with erythroid maturation in each dataset. A hypergeometric test indicated that miRNA target genes have significant overlaps with the “erythroid genes”, suggesting their common and stringent involvement in erythropoiesis (Figure 8F and Supplementary information, Figure S9G). Taken together, our findings suggest that these QKI5-responsive miRNAs may have fundamentally erythroid-regulatory activities.
Discussion
The impact of miRNAs on gene expression clearly correlates with their abundance, and miRNA abundance can be regulated in seemingly endless ways1. In the last decade, identification of the pathways by which miRNAs are produced, matured and turned over has revealed many aspects of their biogenesis. Microprocessor-mediated processing is the first step of cleavage, and its efficiency is crucial for determining miRNA abundance. Interestingly, pri-miRNA processing is also regulated during processes such as differentiation and tumor progression. For example, during early development, numerous pri-miRNAs are expressed but are not efficiently converted into mature miRNAs30. Similarly, reduced processing contributes to widespread downregulation of miRNAs in many human cancers31,32,33. These findings have uncovered a significant role for the regulated Microprocessor-mediated mechanism of miRNA biogenesis. Determinants around miRNA stem loops are better understood11,12,34, as illustrated by both the design and prediction35 of Drosha substrates. However, other cis-elements that outside of the typical pri-miRNA stem loops, are relatively little known. In fact, compared with the small size of stem loops, pri-miRNAs contain more distant recognition elements that may enable specificity concerning when, where and to what extent they are expressed. These elements are attractive subjects for further studies in which additional unique determinants used by different pri-miRNAs would be identified.
Our present study revealed that a distal cis-element within pri-124-1 could dynamically regulate its processing by Microprocessor (Figure 8G). This QRE motif is distinguished from the previously reported stem-loop proximal elements by being able to act at a distance. The mechanism by which the QRE motif affects Microprocessor processing is through a proposed RNA-RNA interaction. In fact, despite being single-stranded, RNA molecules often fold back on themselves to form base-paired segments between short stretches of complementary sequences; therefore, sites separated by many base pairs may not actually be as far apart as previously thought. In the case of pri-124-1, two complementary regions (R1 and R2) near the QRE and pri-124-1 stem loops interact with each other and bring the QKI5-recruited Microprocessor into close proximity to pri-124-1 stem loops. A recent report also illustrated that two complementary regions within pri-17∼92 could selectively inhibit its processing by Microprocessor36. Thus, the local RNA conformations formed through complementary cis-regulatory regions would contribute a lot to pri-miRNA processing. Moreover, the QRE motif also functions as a “Microprocessing enhancer” through the formation of a specific RNA structure, which resembles a transcription enhancer that activates the promoter through special chromatin structures. Yet two previous studies have reported the contradictory results regarding the effect of QKI isoforms on miRNA expression; Chen et al.19 indicated the association of mature miR-20 with and its stabilization by either QKI isoforms, whereas Wang et al.20 showed prevention of pri-miR-7 processing by QKI5 and QKI6 through their binding near the stem-loop structure. However, both of these studies failed to illustrate the detailed mechanisms underlining QKI regulation. Our finding differs from theirs as we established the QKI5-directed recruitment mechanism of Microprocessor on pri-miRNAs, and this regulation is mediated by a distant RNA motif far from the central stem-loop structures.
Our identification of the functional QRE motif may represent an emerging model where pri-miRNA recognition is a modular phenomenon in which each module contributes modestly, and each pri-miRNA depends on individual modules to varying degrees. As pri-miRNAs can be very long, numerous RBP motifs either adjacent or distant to the processing site could act synergistically to regulate pri-miRNA processing. This synergy is critical for signal integration by different RBPs, e.g., in pri-124-1 processing, we also identified a previously reported protein, KSRP, as a potential negative regulator (Figure 2B). Thus, QKI5 and KSRP could transmit separate signals to Microprocessor complex. A previous study reported that only 25% of the fluctuations in miRNA expression in developing B cells could be attributed to changes in transcription as inferred by the pri-miRNA abundance in the same cells37; thus post-transcriptional events would bring more specificity to the developmental regulation of miRNA biogenesis.
Finally, we identified a unique miRNA signature subjected to QKI5 regulation during erythroid differentiation. Functional screening of these miRNAs in erythroid differentiation further verified their repressive roles (miR-124, miR-454, miR-130b and miR-135a). Thus, the downregulation of QKI5 along with erythroid differentiation inhibits the processing of a subset of antagonistic miRNAs, which may become a common pattern to render those miRNAs harmless at the post-transcriptional level. This new paradigm for QKI5 allows more inputs on QKI5-associated gene regulation, i.e., in addition to its characterized roles in regulating pre-mRNA splicing, circular RNA formation, mRNA export, translation and stability24,38,39,40,41, QKI5 also acts as a component of Microprocessor to affect pri-miRNA processing pathway.
The study of regulators and their accessory factors responsible for miRNA biogenesis will help determine the sophisticated mechanisms that fine-tune the miRNA machinery. Further investigations should unravel the cooperation of multiple mechanisms to coordinate the exquisite outputs of certain miRNAs at the post-transcriptional level. Such knowledge will not only expand our understanding of how miRNA abundance is controlled under physiological conditions but also will present exciting opportunities for screening of compounds manipulating the miRNA machinery that can be used in therapeutic applications.
Materials and Methods
Cell culture
Human umbilical cord blood (UCB) was obtained from normal full-term deliveries after informed consent as approved by the Research Ethics Committee of Peking Union Hospital (Beijing, China). Mononuclear cell (MNC) fractions were isolated from UCB by Percoll density gradient (d = 1.077; Amersham Biotech, Germany). CD34+ cells were enriched from MNCs through positive immunomagnetic selection (CD34 MultiSort kit, Miltenyi Biotec, Bergisch-Glad-bach, Germany). The isolated CD34+ hematopoietic progenitor cells (HPCs) were cultured in IMDM supplemented with 30% fetal bovine serum (FBS; Hyclone), 1% BSA, 100 μM 2-ME, 2 ng/ml recombinant human IL-3, 100 ng/ml recombinant human SCF (Stem Cell Technologies, Vancouver, BC, Canada), 2 U/ml recombinant human EPO (R&D Systems, Minneapolis, MN, USA), 60 mg/ml penicillin and 100 mg/ml streptomycin. Cells were harvested every 3-5 days. Human erythroleukemia cell lines K562 and HEL were maintained in RPMI1640 supplemented with 10% FBS (Gibco, Carlsbad, CA, USA). Erythroid differentiation of K562 and HEL cells was induced using 30 μM hemin (Sigma-Aldrich, Deisenhofen, Germany) for 0, 24, 48 and 72 h. 293T cells were obtained from American Type Culture Collection and grown in DMEM with 10% FBS. For morphological analysis, cells were smeared on glass slides by centrifugation, stained with May-Grünwald/Giemsa and analyzed at 400× magnification under a microscope (Nikon TE2000) equipped with a digital camera.
Oligonucleotides and constructs
MiR-124 mimics, miRNA inhibitors (anti-124) and negative control molecules (NC) were obtained from Dharmacon (Austin, TX, USA) and transfected with DharmFECT1 (Dharmacon, Austin, TX, USA) at a final concentration of 60 nM. siRNA smart pools (specifically for QKI5 and RBPs) and control siRNAs (Si-Con) were synthesized by Dharmacon and transfected (100 nM) using DharmFECT1. For QKI5 overexpression, the human QKI5 (NM_006775) cDNA ORF Clone was purchased from Origene (pCMV6-QKI5 in Figures 2H, 5D and 5K, and QKI5 in Figure 3B, RC205779). Mutations (QKI5V157E) or deletions (QKI5ΔKH) on the KH domain of QKI5 cDNA were created using the QuickChange Site-Directed Mutagenesis kit (Stratagene, CA, USA). For pri-124-1 overexpression, a 579-bp wild-type (124_WT) construct containing QRE and pri-124-1 stem loops from human genomic DNA was cloned into pCMV6 vector downstream of the CMV promoter. The QRE mutant pri-124-1 construct (124_MUT) was created using the QuickChange Site-Directed Mutagenesis kit. The mutant sequence was displayed in Figure 6D and also shown in Supplementary information, Table S4. Pri-124-1 variants, which have different RNA-RNA interaction mutant regions (124-R1_MUT, 124-R2_MUT and 124-R1+R2_MUT) were also created using the QuickChange Site-Directed Mutagenesis kit. For RNase H protection assay, a segment containing 1-174 nt (124-QRE, containing the QRE motif and miR-124 stem loop) of 124_WT was subcloned from 124_WT. For the QRE-introduced pri-23a∼27a∼24-2 constructs, QRE-containing oligonucleotides (5′-GGGTAATTAACACGGGGGA-3′) were inserted ∼60 bp upstream of miR-23a stem loop or ∼30 bp downstream of the miR-24-2 stem loop. These QRE-containing pri-23a∼27a∼24-2 constructs were cloned into the pCMV6 vector to generate 23a_QREup or 23a_QREdown, respectively. For the reporter gene assay of miR-124 targets, the reverse complementary sequence to miR-124 was inserted into pGL-3 downstream of the firefly luciferase gene to generate a positive reporter construct (positive). The 3′ UTR of human c-MYB and TAL1 mRNA was PCR amplified and cloned into pGL3 downstream of the firefly luciferase gene to generate the wild-type reporters (c-MYB and TAL1). Mutations of the miR-124 binding sites in the c-MYB and TAL1 mRNA sequences were created using the QuikChange Site-Directed Mutagenesis kit. Transfection of these constructs was carried out with either Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) for 293T cells or Lipofectamine LTX & PLUS for K562 and HEL cells according to the manufacturer's protocols. All primers are listed in Supplementary information, Table S4.
Lentivirus production
For miR-124 overexpression, a same 579-bp construct containing QRE and pri-124-1 stem loops from human genome was cloned into a pol II promoter lentivector (pCDH-CMV-MCS-EF1, CD511B-1, System Biosciences, CA, USA) to generate lenti-124, or an 80-bp pri-124-1 stem loop was cloned downstream of a pol III promoter lentivector (pSIH1, SI501A-1, System Biosciences, CA, USA) to generate pSIH-124. For miR-124 inhibition, miRZips lentiviral vector (lenti-Zip-124) was purchased from System Biosciences (MZIP124-PA-1). For QKI5 overexpression, the QKI5 cDNA was cloned downstream of the CMV pol II promoter (pWPXL, Addgene) to generate lenti-QKI5. For QKI5 knockdown, the double-stranded siRNA templates specific to QKI5 were cloned into lentivector pSIH1 to generate lenti-si_QKI5. The matching lentivirus packaging kit was purchased from System Biosciences (SBI, CA, USA) and used according to the manufacturer's instructions. The harvested viral particles (lenti-124, lenti-Zip-124, lenti-QKI5 and lenti-si_QKI5) were added to HPCs or K562 cells, respectively. The cells were washed the next day with PBS and plated for subsequent experiments.
RNA isolation and quantitative real-time PCR
Total RNA was extracted from cells using Trizol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instruction. The RNA was quantified by absorbance at 260 nm. cDNA was synthesized by M-MLV reverse transcriptase (Invitrogen) from 2 μg of total RNA. Oligo (dT) 18 primers were used as the RT primers for reverse transcription of mRNAs and pri-miRNAs. Stem-loop RT primers were used as the RT primers for reverse transcription of mature miRNAs. q-PCR was carried out in the BIORAD IQ5 real-time PCR System (Biorad, Foster City, CA, USA) using SYBR Premix Ex Taq kit (Takara, Dalian, China) according to the manufacturer's instructions. For mRNAs and pri-miRNAs, the data were normalized using endogenous GAPDH as a control. For the measurement of mature miRNAs, the data were normalized using the endogenous U6 snRNA control. Quantitative RNA-IP analysis was performed using 1 μl template RNA in 20 μl reverse-transcription reactions. The data were presented as the fold change or enrichment of precipitated RNA associated with QKI5 relative to input RNAs. The comparative Cq method was used to quantify the target genes relative to an endogenous control. For each individual analysis, one of the samples was designated as the calibrator and given a relative value of 1.0. All quantities were expressed as n-fold relative to the calibrator. The primers used for PCR are listed in Supplementary information, Table S4.
Northern blot analysis
For pri-124-1 hybridization, 30 μg of total RNAs collected from hemin-treated K562 and HEL cells, or 60 μg of total RNAs collected from EPO-treated HPCs were resolved on 3.5% PAGE gels, and northern blot was performed according to the manufacturer's protocol. Digoxigenin-labeled antisense pri-124-1 probes were prepared using T7 RNA polymerases by in vitro transcription with the DIG Northern Starter Kit (Roche, NJ, USA). For miR-124 hybridization, 40 μg of total RNAs collected from hemin-treated K562 and HEL cells, or 60 μg of total RNAs collected from EPO-treated HPCs were isolated on 15% PAGE gels, and northern blot was performed as previously described42. Isotope-labeled antisense miR-124 probes were prepared using T4 Polynucleotide Kinase. The sequences of the oligonucleotide probes are listed in Supplementary information, Table S4.
RACE analysis
To isolate the full-length pri-124-1, 5′ and 3′ RACE reactions were performed on total RNA of 48 h hemin-treated K562 cells using the 5′-Full RACE Kit and 3′-Full RACE Kit (TaKaRa, Dalian, China) according to the manufacturer's protocol. Primers used for the RACE experiment are listed in Supplementary information, Table S4.
Luciferase reporter assay
For functional analysis of the miR-124-1 promoter, 293T cells were co-transfected with 0.4 μg of pGL-3 basic constructs containing the miR-124-1 promoter, along with 0.02 μg of the pRL-TK control vector and 0.2 μg of the pcDNA3.1 constructs with full-length cDNA of QKI5 (pCMV6-QKI5) and their corresponding empty vector controls. For miRNA target analysis, 293T cells were co-transfected with 0.4 μg of the reporter construct, 0.02 μg of the pRL-TK control vector and 5 pmol of miR-124 mimic or negative control (scramble). Cells were harvested 48 h post transfection and assayed with Dual Luciferase Assay (Promega, WI, USA) according to the manufacturer's instructions. All transfection assays were carried out in triplicate.
Western blot analysis
Whole-cell lysate or nuclear extract was subjected to western blot analysis following a previously described procedure43. The following antibodies were used for western blotting: GAPDH (0411) was purchased from Santa Cruz Biotechnology. QKI5 (AB9904) was purchased from Millipore Company. KSRP (ab95229), ELAVL1 (ab28660), EIF4B (ab38359), SRSF10 (ab112054), PABPC1 (ab21060), Drosha (ab12286), DGCR8 (ab36865), DDX17 (ab66764), FUS (ab23439), C-MYB (ab62824), TAL1 (ab119754), GST (ab6613) and Flag (ab1162) were purchased from Abcam Company. All the experiments were repeated at least once with similar results. Immunoblots were quantified using ImageJ software.
RNA-protein complex immunoprecipitation
1 × 107 K562 or HEL cells were harvested in ice-cold PBS. Next, the cell pellets were resuspended in 1 ml lysis buffer B (50 mM Tris, pH 7.4, 150 mM NaCl, 0.05% Igepal, 1 mM PMSF, 1 mM aprotinin, 1 mM leupeptin and 2 mM VRC) and subjected to two rounds of gentle sonication. After the lysates were centrifuged at 12 000 rpm for 15 min, the supernatants were precleared with Dynabeads (Invitrogen) in lysis buffer B with a supplement of 10 μg yeast tRNA (Sigma). Then, the precleared lysates were used for RIP with either anti-QKI5 or a rabbit isotype control immunoglobin G (IgG). RNA-IP was carried out for 4 h at 4 °C. The beads were washed three times with the same lysis buffer B, with the last wash containing an additional 0.5% sodium deoxycholate, followed by extraction with buffer C (100 mM Tris, pH 6.8, 4% SDS, 12% β-mercaptoethanol and 20% glycerol) at room temperature for 10 min. The IP material was used for RNA extraction with Trizol. For RT-PCR, each RNA sample was treated with DNase I (Ambion, DNA-freeTM kit); then, reverse transcription was performed with random primers. The resulting material was used for PCR analyses. The fold enrichments of RNAs by QKI5 antibodies over IgG were assayed by q-PCR. The primers used for RNA-IP-PCR experiment are listed in Supplementary information, Table S4.
RNA EMSA
The biotin-labeled miR-124-QRE and p27-QRE RNA probe, cold probe and mutant probe were synthesized by RiboBio Life Science Company. A total of 50 fmol of biotin-labeled RNA probes were incubated with various concentrations of Flag-QKI5 (purified from 293T cells by anti-Flag M2 magnetic beads, Sigma-Aldrich) using the LightShift Chemiluminescent RNA EMSA Kit (Pierce, IL, USA) according to the manufacturer's protocol. Competition experiments were performed with 200-fold molar excess of the unlabeled probe (cold probe) preincubation. The reactions were incubated at room temperature for 30 min before adding RNA loading dye and separated by native 8% PAGE. The probes used for the RNA EMSA experiment are listed in Supplementary information, Table S4.
Protein-RNA complex immunoprecipitation by RNA pull-down
MBP-affinity purification was used to identify proteins that associated with pri-miRNAs. The expression plasmid MS2-MBP was a gift from Dr Lingling Chen (Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences (CAS)), and MS2-MBP was expressed and purified from E. coli following a protocol from the Steitz lab. Three bacteriophage MS2 coat protein-binding sites (5′-cgtacaccatcagggtacgagctagcccatggcgtacaccatcagggtacgactagtagatctcgtacaccatcagggtacg-3′) were inserted downstream of pri-124-1 or pri-23a∼27a∼24-2 using Stratagene QuikChange Site-Directed Mutagenesis Kit. To obtain proteins associated with pri-miRNAs, 293T cells were transfected with MS2-tagged pri-miRNA constructs, and 10 million cells were used for each immunoprecipitation assay. The cells were harvested 48 h post transfection and subjected to RNA pull-down analysis as described elsewhere44.
Affinity purification of K562 and HEL cell extracts
HEK293T cells were cultured in 10-cm dishes and transfected with Flag-QKI5 or Flag-RFP constructs. Forty-eight hours post transfection, the cells were lysed, and Flag-tagged bait proteins were recovered on Flag-agarose beads by incubating at 4 °C for 2 h. A total of 1 × 108 of K562 and HEL cells were lysed in 2 ml lysis buffer and precleared with Flag-agarose beads. The supernatant was incubated with Flag-agarose beads that were prebound with bait proteins at 4 °C overnight. The bound proteins were eluted, boiled, subjected to SDS-PAGE, and immunoblotted with antibodies against Drosha and DGCR8.
FPLC chromatography
K562 NEs were applied to a Superose 6 gel filtration column (GE Healthcare Life Sciences) that had been equilibrated with dithiothreitol-containing buffer and calibrated with protein standards (blue dextran, 2 000 kDa; thyroglobulin, 669 kDa; ferritin, 440 kDa; bovine serumalbumin, 67 kDa; and RNase A, 13.7 kDa; all from GE Healthcare Life Sciences). The column was eluted at a flow rate of 0.5 ml/min, and fractions were collected.
Protein-protein complex immunoprecipitation
K562 and HEL cellular extracts were incubated with QKI5 mouse monoclonal antibody or normal mouse IgG at 4 °C overnight, followed by the addition of protein A/G-Agarose (Roche) for 2 h at 4 °C. The beads were then washed, and the immune complexes were subjected to SDS-PAGE followed by immunoblotting with antibodies against Drosha and DGCR8. Immunodetection was performed using enhanced chemiluminescence (Engreen) according to the manufacturer's instructions.
GST pull-down
GST fusion constructs were expressed in BL21 E. coli cells, and crude bacterial lysates were prepared by sonication in PBS buffer in the presence of protease inhibitor cocktails. In GST pull-down assays, ∼5 μg of purified GST fusion protein was mixed with 293T cell lysates expressing Flag-Drosha, Flag-DGCR8 or its deletion mutants and incubated in NETN buffer (20 mM Tris-HCl, pH 8.0, 100 mM NaCl, 0.5% NP-40, 1 mM EDTA and the protease inhibitor cocktails) at 4 °C for 1 h. The binding reaction was then added to 30 μl of glutathione-Sepharose beads and mixed at 4 °C for 2 h. The beads were washed three times with NETN buffer, resuspended in 30 μl of 1× SDS-PAGE loading buffer, resolved on 10% gels and immunoblotted with antibodies against Flag and GST.
Immunofluorescence
K562, HEL and HEK293T cells were grown on cover slips and fixed with 4% (v/v) paraformaldehyde for 15 min at room temperature. The cells were then permeabilized with 0.5% Triton X-100 for 10 min and incubated with 5% (v/v) goat serum for 30 min at room temperature followed by incubation with diluted primary antibody at 4 °C overnight. The cells were then probed with Alexa Fluor 488 and 594 secondary antibodies (Invitrogen). Fluorescence signals were detected using a confocal fluorescence microscope (NikonEZ-C1, Nikon).
In vitro processing assays
Processing reactions were performed as described previously17. Briefly, Hela cell nuclear extracts (Hela NE, Cil Biotech, Belgium) was treated with QKI5 antibody-coated protein G beads at 4 °C for 30 min (refer to as “QKI5-depleted Hela NE”). In 30 μl processing reaction, 6.4 mM MgCl2, 1 unit/ml of Ribonuclease Inhibitor (Invitrogen), the 32P-labeled transcripts of 1 × 105 cpm, and 20 μl of QKI5-depleted Hela NE, were incubated with different concentrations of Flag-QKI5 protein (1×, 10 ng/μl; 2×, 20 ng/μl; 4×, 40 ng/μl) purified from 293T cells by anti-Flag M2 magnetic beads (Sigma-Aldrich) or not. The reaction mixture was incubated at 37 °C for 120 min. The 32P-labeled RNA substrates were prepared by standard in vitro transcription with T7 RNA polymerase in the presence of [α-32P]-UTP using 124_WT, 124_MUT, 23a, 124-R1_MUT, 124-R2_MUT and 124-R1+R2_MUT constructs as templates. RNA was phenol-extracted from the reaction mixture and analyzed on 8% denaturing urea-polyacrylamide gel.
RNase H protection assay
A standard RNase H protection reaction is carried out as described previously11,45,46. Briefly, in 25 μl reaction, 32P-labeled 124-QRE transcripts of 1 × 105 cpm, 10 μg/ml DNA oligonucleotide, 12 mM HEPES, pH 8.0, 60 mM KCl, 3 mM MgCl2, 1 mM DTT, 20 U of RNasin (Invitrogen) and 1 U of E. coli RNase H (TaKaRa, 2150A) were incubated with 10 ng/μl Flag-QKI5 protein purified from 293T cells by anti-Flag M2 magnetic beads (Sigma) or not. The reaction mixture was incubated at 30 °C for 40 min. The 32P-labeled 124-QRE substrates were prepared by standard in vitro transcription with T7 RNA polymerase in the presence of [α-32P]-UTP using 124-QRE construct as template. RNA was phenol-extracted from the reaction mixture and analyzed on 8% denaturing urea-polyacrylamide gel.
Gel shift experiments
RNA band shift experiments were performed as described previously47,48. Briefly, the biotin-labeled R2_WT, biotin-labeled R2_MUT, R1_WT and R1_MUT fragments were synthesized in vitro (RiboBio). RNA band shift experiments were performed in 1× structure buffer (Ambion) in a total reaction mixture volume of 15 μl as follows: 125 nM of biotin-labeled R2_WT or biotin-labeled R2_MUT, and different concentrations of unlabeled R1_WT or R1_MUT fragments at 30 °C for 20 min. Prior to gel loading, the binding reactions were mixed in 4.5 μl of native loading dye (50% glycerol, 0.5× TBE, 0.1% bromophenol blue and 0.1% xylenecyanol) and run on native 8% polyacrylamide gels. The RNAs used for this experiment are listed in Supplementary information, Table S4.
Flow cytometry
The cells were harvested at the indicated time points and washed twice at 4 °C in PBS/0.5% BSA to block Fc receptors. The transduced CD34+ HPCs were assessed for green fluorescence (GFP) and CD235a expression after staining with PE-conjugated anti-CD235a and APC-conjugated anti-CD71 antibodies (BD Biosciences PharMingen). Mouse BM cells were stained with PE-conjugated anti-33 antibodies (eBioscience, CA, USA). Flow cytometry was carried out on a C6 Flow Cytometer Instrument (BD Biosciences, Franklin Lakes, NJ, USA).
Mice and transplantation assays
All animal experiments were performed with the approval of the Research Ethics Committee of Peking Union Medicine College. Purified human CD34+ HPCs were exposed to lentiviruses expressing GFP, QKI5 or miR-124 at MOI of 15. All cells were incubated for 16 h at 37 °C in 5% CO2 before being washed twice in PBS. A small number of cells from each transduction group were cultured in vitro, whereas the remaining cells were transplanted into mice. NOD/SCID mice of 5 weeks of age were each injected with 1 × 107 transduced cells in the lateral tail veins. These mice had been conditioned with 2× 450 cGy γ-radiation from a 137Cs source 4 h previously. Mice were killed 4 weeks post transplantation, and the BM and SP were harvested and processed into single-cell suspensions and used in subsequent experiments.
Small RNA-seq
K562 cells were infected with lentiviruses expressing QKI5 or a GFP control and sorted by FACS to collect the GFP-positive cells (Supplementary information, Figure S7A). Small RNA was isolated from the total RNA with the mirVana miRNA Isolation Kit (Life Technologies, CA, USA) following the manufacturer's instructions. Small RNA libraries were prepared with the Ion Total RNA-Seq Kit v2 (Life Technologies) following the manufacturer's instructions and then were sequenced on the Ion Torrent Proton System using Ion PI Chips with two libraries (Con: K562-lenti-GFP; QKI5: K562-lenti-QKI5) sequenced on one chip. Roughly, the libraries were sized using the Experion system (BioRad, Hercules, CA, USA) and normalized to a working concentration of 10 pM. The libraries were then diluted to 1 pM, and Ion sphere particle emulsion PCR amplification was completed on an Ion OneTouch 2 instrument according to the recommendations in the Ion PI Template OT2 200 Kit (Life Technologies, Carlsbad, CA, USA). After emulsion PCR, a Qubit Ion sphere quality control assay was performed to verify that the percentage of templated ion sphere particles (ISPs) was in the optimal range of 10%-25% prior to enrichment and sequencing. The loaded chips were sequenced with the Ion PI Sequencing 200 kit. The sequence data were filtered, the expression profiles were constructed and the adapter dimer reads were separated from the miRNA reads already identified in miRBase 17.0. A full list of relative miRNA expression is listed in Supplementary information, Table S6.
Bioinformatics analysis of miRNA targets
The target genes of four miRNAs (miR-124-3p, miR-454-3p, miR-130b-3p and miR-135a-5p) were downloaded from miRTarBase49 and starBase50. To retrieve relatively reliable miRNA target genes, it should be noted that the target genes with the annotation “Weak” interactions in miRTarBase were excluded, and those target genes supported by at least five experiments and validated by at least four prediction tools in starBase were adopted. Gene Ontology analysis of the target genes was performed by the functional annotation tool DAVID51 with the default parameter setting.
To further validate the essential role of the miRNA target genes in erythropoiesis, a total of four erythropoiesis-associated RNA-seq or array datasets (GSE43023, GSE53635, GSE61566 and GSE53983) were downloaded from GEO database. For each dataset the top 10% expressed genes at each stage of erythropoiesis were defined as the erythropoiesis-associated highly expressed genes, and the upregulated genes during erythropoiesis were identified as a fold change > 1.5 between any pairwise comparisons and an RPKM > 0.1 in most of the differentiation stages. The “Erythroid genes” were defined as the overlap between highly expressed genes and upregulated genes for each dataset.
Statistical analysis
For all quantified data, the mean ± SEM is presented. Statistical significance between two experimental groups is indicated by an asterisk, and comparisons were made using Student's t-test. P-values < 0.05 were considered significant.
Data deposition
The GEO accession numbers for microarray and small RNA-seq analyses are GSE30380 and GSE57363.
Author Contributions
J Yu and F Wang conceived and supervised the study. F Wang, W Song and H-M Zhao performed most of the experiments. Y-N Ma, D Zhai and J-N Pi performed the in vitro processing assay. Y-X Li, Y-M Si and R Su performed cell culture analysis. J-Y Xu and L Dong performed bioinformatics analysis. M-M Zhang, X-X Ren, F Miao and W-J Liu performed GST pull-down analysis. Y Zhu and F Li are involved in mouse experiments. J-W Zhang, A-B He, G Shan, J-Y Hui and L-F Wang provided suggestions to this work. J Yu and F Wang analyzed the data and wrote the paper.
Competing Financial Interests
The authors declare no competing financial interests.
References
Ameres SL, Zamore PD . Diversifying microRNA sequence and function. Nat Rev Mol Cell Biol 2013; 14:475–488.
Cech TR, Steitz JA . The noncoding RNA revolution-trashing old rules to forge new ones. Cell 2014; 157:77–94.
Yang JS, Lai EC . Alternative miRNA biogenesis pathways and the interpretation of core miRNA pathway mutants. Mol Cell 2011; 43:892–903.
Ha M, Kim VN . Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 2014; 15:509–524.
Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ . Processing of primary microRNAs by the microprocessor complex. Nature 2004; 432:231–235.
Gregory RI, Yan KP, Amuthan G, et al. The microprocessor complex mediates the genesis of microRNAs. Nature 2004; 432:235–240.
Nguyen TA, Jo MH, Choi YG, et al. Functional anatomy of the human microprocessor. Cell 2015; 161:1374–1387.
Bartel DP . MicroRNAs: target recognition and regulatory functions. Cell 2009; 136:215–233.
Shukla GC, Singh J, Barik S . MicroRNAs: processing, maturation, target recognition and regulatory functions. Mol Cell Pharmacol 2011; 3:83–92.
Czech B, Hannon GJ . Small RNA sorting: matchmaking for Argonautes. Nat Rev Genet 2011; 12:19–31.
Han J, Lee Y, Yeom KH, et al. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell 2006; 125:887–901.
Auyeung VC, Ulitsky I, McGeary SE, Bartel DP . Beyond secondary structure: primary-sequence determinants license pri-miRNA hairpins for processing. Cell 2013; 152:844–858.
Guil S, Caceres JF . The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat Struct Mol Biol 2007; 14:591–596.
Trabucchi M, Briata P, Garcia-Mayoral M, et al. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature 2009; 459:1010–1014.
Chang HM, Triboulet R, Thornton JE, Gregory RI . A role for the Perlman syndrome exonuclease Dis3l2 in the Lin28-let-7 pathway. Nature 2013; 497:244–248.
Heo I, Ha M, Lim J, et al. Mono-uridylation of pre-microRNA as a key step in the biogenesis of group II let-7 microRNAs. Cell 2012; 151:521–532.
Heo I, Joo C, Cho J, Ha M, Han J, Kim VN . Lin28 mediates the terminal uridylation of let-7 precursor microRNA. Mol Cell 2008; 32:276–284.
Fang W, Bartel DP . The menu of features that define primary microRNAs and enable de novo design of microRNA genes. Mol Cell 2015; 60:131–145.
Chen AJ, Paik JH, Zhang H, et al. STAR RNA-binding protein Quaking suppresses cancer via stabilization of specific miRNA. Genes Dev 2012; 26:1459–1472.
Wang Y, Vogel G, Yu Z, Richard S . The QKI-5 and QKI-6 RNA binding proteins regulate the expression of microRNA 7 in glial cells. Mol Cell Biol 2013; 33:1233–1243.
Hafner M, Landthaler M, Burger L, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 2010; 141:129–141.
Teplova M, Hafner M, Teplov D, Essig K, Tuschl T, Patel DJ . Structure-function studies of STAR family Quaking proteins bound to their in vivo RNA target sites. Genes Dev 2013; 27:928–940.
Zhao L, Ku L, Chen Y, Xia M, LoPresti P, Feng Y . QKI binds MAP1B mRNA and enhances MAP1B expression during oligodendrocyte development. Mol Biol Cell 2006; 17:4179–4186.
Larocque D, Galarneau A, Liu HN, Scott M, Almazan G, Richard S . Protection of p27(Kip1) mRNA by quaking RNA binding proteins promotes oligodendrocyte differentiation. Nat Neurosci 2005; 8:27–33.
Wright PR, Georg J, Mann M, et al. CopraRNA and IntaRNA: predicting small RNA targets, networks and interaction domains. Nucleic Acids Res 2014; 42:W119–W123.
Larocque D, Richard S . QUAKING KH domain proteins as regulators of glial cell fate and myelination. RNA Biol 2005; 2:37–40.
Zhao L, Mandler MD, Yi H, Feng Y . Quaking I controls a unique cytoplasmic pathway that regulates alternative splicing of myelin-associated glycoprotein. Proc Natl Acad Sci USA 2010; 107:19061–19066.
Li Y, Deng C, Hu X, et al. Dynamic interaction between TAL1 oncoprotein and LSD1 regulates TAL1 function in hematopoiesis and leukemogenesis. Oncogene 2012; 31:5007–5018.
Vegiopoulos A, Garcia P, Emambokus N, Frampton J . Coordination of erythropoiesis by the transcription factor c-Myb. Blood 2006; 107:4703–4710.
Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, Hammond SM . Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev 2006; 20:2202–2207.
Garzon R, Fabbri M, Cimmino A, Calin GA, Croce CM . MicroRNA expression and function in cancer. Trends Mol Med 2006; 12:580–587.
Lu J, Qian J, Chen F, Tang X, Li C, Cardoso WV . Differential expression of components of the microRNA machinery during mouse organogenesis. Biochem Biophys Res Commun 2005; 334:319–323.
Xia H, Yamada S, Aoyama M, et al. Prognostic impact of microRNA-145 down-regulation in adult T-cell leukemia/lymphoma. Hum Pathol 2014; 45:1192–1198.
Yeom KH, Lee Y, Han J, Suh MR, Kim VN . Characterization of DGCR8/Pasha, the essential cofactor for Drosha in primary miRNA processing. Nucleic Acids Res 2006; 34:4622–4629.
Helvik SA . Snove O Jr, Saetrom P . Reliable prediction of Drosha processing sites improves microRNA gene prediction. Bioinformatics 2007; 23:142–149.
Du P, Wang L, Sliz P, Gregory RI . A biogenesis step upstream of microprocessor controls miR-17∼92 expression. Cell 2015; 162:885–899.
Kuchen S, Resch W, Yamane A, et al. Regulation of microRNA expression and abundance during lymphopoiesis. Immunity 2010; 32:828–839.
Conn SJ, Pillman KA, Toubia J, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell 2015; 160:1125–1134.
Yang G, Lu X, Wang L, et al. E2F1 and RNA binding protein QKI comprise a negative feedback in the cell cycle regulation. Cell Cycle 2011; 10:2703–2713.
Zhang Y, Lu Z, Ku L, Chen Y, Wang H, Feng Y . Tyrosine phosphorylation of QKI mediates developmental signals to regulate mRNA metabolism. EMBO J 2003; 22:1801–1810.
Zong FY, Fu X, Wei WJ, et al. The RNA-binding protein QKI suppresses cancer-associated aberrant splicing. PLoS Genet 2014; 10:e1004289.
Yu J, Wang F, Yang GH, et al. Human microRNA clusters: genomic organization and expression profile in leukemia cell lines. Biochem Biophys Res Commun 2006; 349:59–68.
Yu J, Ryan DG, Getsios S, Oliveira-Fernandes M, Fatima A, Lavker RM . MicroRNA-184 antagonizes microRNA-205 to maintain SHIP2 levels in epithelia. Proc Natl Acad Sci USA 2008; 105:19300–19305.
Yin QF, Yang L, Zhang Y, et al. Long noncoding RNAs with snoRNA ends. Mol Cell 2012; 48:219–230.
Gunzl A, Palfi Z, Bindereif A . Analysis of RNA-protein complexes by oligonucleotide-targeted RNase H digestion. Methods 2002; 26:162–169.
Vickers TA, Koo S, Bennett CF, Crooke ST, Dean NM, Baker BF . Efficient reduction of target RNAs by small interfering RNA and RNase H-dependent antisense agents. A comparative analysis. J Biol Chem 2003; 278:7108–7118.
Brantl S, Wagner EG . Antisense RNA-mediated transcriptional attenuation occurs faster than stable antisense/target RNA pairing: an in vitro study of plasmid pIP501. EMBO J 1994; 13:3599–3607.
Overloper A, Kraus A, Gurski R, et al. Two separate modules of the conserved regulatory RNA AbcR1 address multiple target mRNAs in and outside of the translation initiation region. RNA Biol 2014; 11:624–640.
Hsu SD, Tseng YT, Shrestha S, et al. miRTarBase update 2014: an information resource for experimentally validated miRNA-target interactions. Nucleic Acids Res 2014; 42:D78–D85.
Li JH, Liu S, Zhou H, Qu LH, Yang JH . starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res 2014; 42:D92–D97.
Huang da W, Sherman BT, Lempicki RA . Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009; 37:1–13.
Acknowledgements
We are thankful to Lingling Chen (Institute of Biochemistry and Cell Biology, CAS, China) for the MS2-MBP plasmid, and Bin Zhu (Peking Union Hospital, China) for assistance in umbilical cord blood preparation, and Bo Wen (Fudan University, China) for helpful discussions. J Yu lab is supported by the National Key Research and Development Program of China (2016YFA0100601), the National Key Basic Research Program of China (2015CB94300), the National Natural Science Foundation of China (81530007, 9144011, 31371322 and 31471227) and CAMS Initiative for Innovative Medicine (2016-12M-3-002).
Author information
Authors and Affiliations
Corresponding author
Additional information
( Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary information
Supplementary information, Table S1
List of miRNA candidates involved in the initial screening, related to Figure 1B. (PDF 117 kb)
Supplementary information, Table S2
The real-time PCR results of different transcripts of miRNAs, related to Figure 1B. (XLS 32 kb)
Supplementary information, Table S3
RNA binding protein (RBP) that are predicted to bind primary miR-124-1 (from RBPDB database), related to Figure 2. (XLS 93 kb)
Supplementary information, Table S4
Oligo sequences used in this study. (XLS 60 kb)
Supplementary information, Table S5
Microarry results from GSE30380. (XLS 22 kb)
Supplementary information, Table S6
The real-time PCR results of different transcripts of miRNAs, related to Figure 8C. (XLS 23 kb)
Supplementary information, Table S7
The Gene Ontology analysis of individual and overlapping miRNA targets. (XLS 231 kb)
Supplementary information, Figure S1
The post-transcriptional regulation of miR-124-1 during erythroipoiesis. (PDF 693 kb)
Supplementary information, Figure S2
QKI5 regulates pri-124-1 processing in erythroid cells. (PDF 480 kb)
Supplementary information, Figure S3
QKI5 binds to pri-124-1 transcript. (PDF 218 kb)
Supplementary information, Figure S4
The regulation of QKI5 on pri-124-1 processing is dependent on the QRE. (PDF 107 kb)
Supplementary information, Figure S5
QKI5 acts on the processing of QRE-introduced pri-23a∼27a∼24-2 transcripts. (PDF 228 kb)
Supplementary information, Figure S6
QKI5 and miR-124 are negative regulators for HPCs erythroid differentiation. (PDF 448 kb)
Supplementary information, Figure S7
Functional “rescue assay” in HPCs. (PDF 842 kb)
Supplementary information, Figure S8
QKI5 and miR-124 are negative regulators for in vivo erythroid differentiation. (PDF 215 kb)
Supplementary information, Figure S9
The regulation of multiple miRNAs by QKI5 in erythroid cells. (PDF 1136 kb)
Rights and permissions
About this article
Cite this article
Wang, F., Song, W., Zhao, H. et al. The RNA-binding protein QKI5 regulates primary miR-124-1 processing via a distal RNA motif during erythropoiesis. Cell Res 27, 416–439 (2017). https://doi.org/10.1038/cr.2017.26
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cr.2017.26
Keywords
This article is cited by
-
Insight into microRNAs’ involvement in hematopoiesis: current standing point of findings
Stem Cell Research & Therapy (2023)
-
A global screening identifies chromatin-enriched RNA-binding proteins and the transcriptional regulatory activity of QKI5 during monocytic differentiation
Genome Biology (2021)
-
A long noncoding RNA binding to QKI-5 regulates germ cell apoptosis via p38 MAPK signaling pathway
Cell Death & Disease (2019)