Abstract
SIRT6 belongs to the mammalian homologs of Sir2 histone NAD+-dependent deacylase family. In rodents, SIRT6 deficiency leads to aging-associated degeneration of mesodermal tissues. It remains unknown whether human SIRT6 has a direct role in maintaining the homeostasis of mesodermal tissues. To this end, we generated SIRT6 knockout human mesenchymal stem cells (hMSCs) by targeted gene editing. SIRT6-deficient hMSCs exhibited accelerated functional decay, a feature distinct from typical premature cellular senescence. Rather than compromised chromosomal stability, SIRT6-null hMSCs were predominately characterized by dysregulated redox metabolism and increased sensitivity to the oxidative stress. In addition, we found SIRT6 in a protein complex with both nuclear factor erythroid 2-related factor 2 (NRF2) and RNA polymerase II, which was required for the transactivation of NRF2-regulated antioxidant genes, including heme oxygenase 1 (HO-1). Overexpression of HO-1 in SIRT6-null hMSCs rescued premature cellular attrition. Our study uncovers a novel function of SIRT6 in maintaining hMSC homeostasis by serving as a NRF2 coactivator, which represents a new layer of regulation of oxidative stress-associated stem cell decay.
Similar content being viewed by others
Introduction
Stem cell exhaustion is considered as one of the hallmarks of aging1,2. Mesenchymal stem cells (MSCs) are adult stem cells with the potential of differentiation into mesodermal lineages, such as osteoblasts, chondrocytes, and adipocytes3, and play an important role in tissue regeneration4. Aged human and animals exhibit decreased number or diminished proliferative potential of MSCs5,6. Accelerated attrition of MSC pool has been observed in human stem cell and mouse models for human premature aging disorders, including Werner syndrome (WS) and Hutchinson Gilford progeria syndrome7,8,9,10. Transplantation of mesoderm-derived stem cells from young mice increases the lifespan and fitness of progeroid mice11,12. While MSC exhaustion is known as a culprit for aging-associated degeneration of mesodermal tissues1, the underlying genetic pathways employed by human MSCs (hMSCs) to maintain homeostasis have not been fully elucidated.
A number of intracellular and intercellular factors are known to maintain stem cell homeostasis13,14 and one of such factors is reactive oxygen species (ROS)5. ROS is normally maintained at a balanced level by cellular antioxidant systems15. Nuclear factor erythroid 2-related factor 2 (NRF2) is a critical redox sensor and is one of the master regulators of antioxidant responses. NRF2 binds to the antioxidant response elements (AREs) and activates the transcription of a number of antioxidant genes such as heme oxygenase 1 (HO-1) that is known for counteracting ROS16. The decline in NRF2-ARE activity is observed in aged cells, and may account for the oxidative stress-associated tissue degeneration17,18. The key factors regulating NRF2 antioxidant pathway in hMSCs, however, remain to be identified.
SIRT6 is one of the seven mammalian sirtuin homologues of yeast Sir2 longevity protein19. Sirt6-deficient mice suffered from multiple acute premature aging syndromes, which ultimately led to premature death within 1 month after birth20. SIRT6 contains a domain with NAD+-dependent deacetylase activity21,22, which accounts for transcriptional repression of target genes such as NF-κB, HIF1α, and c-JUN by deacetylating histone H3 lysine 9 (H3K9) at the gene promoters23,24,25. In addition, silencing SIRT6 in fibroblasts leads to the accelerated cellular senescence possibly due to attenuated deacetylation of H3K9 at the telomere region26. SIRT6 is also involved in DNA damage repair by deacetylation of H3K56 at the damaged DNA region27,28,29. Recently, evidence has emerged that SIRT6 protects cells from oxidative stress-associated DNA damage21, raising a possibility that SIRT6 could be involved in redox-related cellular homeostasis regulation.
Here we demonstrate that SIRT6 is a positive modulator of NRF2-HO-1 antioxidant pathway in hMSCs, a novel mechanism implicated in safeguarding stem cells from oxidative stress-associated functional decay.
Results
SIRT6-depleted hMSCs exhibit accelerated functional decay
To study the role of SIRT6 in hMSCs, we first generated SIRT6-deficient human embryonic stem cells (hESCs). The exon 1 of SIRT6 gene was removed in hESCs by a transcription activator-like effector nuclease (TALEN)-mediated homologous recombination strategy30,31,32 (Figure 1A). Biallelic loss of SIRT6 in SIRT6−/− hESCs was confirmed by genomic PCR and Southern blotting (Supplementary information, Figure S1A and S1B). Loss of mRNA and protein in SIRT6−/− hESCs was verified by reverse transcription-PCR (RT-PCR) and western blotting, respectively (Figure 1B). Immunofluorescence staining demonstrated a punctate staining pattern of SIRT6 in the nuclei of wild-type (WT) hESCs, which was absent in SIRT6−/− hESCs (Figure 1C). The SIRT6-deficient hESCs were cultured for more than 50 passages without discernible morphological abnormality and still exhibited defining characteristics of pluripotency, including the expression of pluripotency markers and the in vivo differentiation potential towards three germ layer lineages (Supplementary information, Figure S1C and S1D).
SIRT6-deficient hMSCs exhibit accelerated cell attrition. (A) Schematic representation of deletion of SIRT6 by removing exon 1 of SIRT6 gene via TALEN-based gene targeting technique. The donor vector contains a neomycin-resistant cassette (neo) allowing for positive selection, and the neo cassette was then removed from the SIRT6 gene locus. (B) Left panel: western blotting analysis of SIRT6 protein in hESCs. Protein extracts from wild-type (WT, SIRT6+/+) and SIRT6-deficient (SIRT6−/−) hESCs were analyzed by western blotting using an anti-SIRT6 antibody. β-actin was used as the loading control. Right panel: RT-PCR analysis of SIRT6 mRNA in hESCs. A pair of PCR primers spanning the junction region of SIRT6 mRNA exon 1 and exon 2 was used. 18S rRNA was used as the loading control. (C) Bright-field and SIRT6 immunofluorescence micrographs of WT and SIRT6-deficient hESCs. DNA was stained by Hoechst 33342. Bright-field scale bar, 200 μm; immunofluorescence scale bar, 20 μm; zoom-field immunofluorescence scale bar, 10 μm. (D) Bright-field micrographs and FACS analysis of the surface markers CD105, CD73, and CD90 in WT and SIRT6-deficient hMSCs. Scale bar, 100 μm. (E) Western blotting analysis of SIRT6 protein in hMSCs. Protein extracts from WT and SIRT6-deficient hMSCs were analyzed by western blotting using anti-SIRT6 antibody. β-actin was used as the loading control. (F) Immunofluorescence analysis showing the absence of SIRT6 protein in the nuclei of SIRT6-deficient hMSCs. Scale bar, 10 μm. (G) SA-β-GAL staining from passage 6-8 showing an accelerated senescence in SIRT6-deficient hMSCs. Percentages of SA-β-GAL-positive cells were calculated. Data were presented as mean ± SEM, n = 5, NS, not significant, **P < 0.01. (H) Western blotting analysis of P16 and P21 protein in hMSCs. Protein extracts from WT and SIRT6-deficient hMSCs at late passage (LP, passage 9) were analyzed by western blotting. β-actin was used as the loading control. (I) Analysis of luciferase activity in the TA muscles of immunodeficient mice by in vivo imaging system (IVIS) demonstrating premature attrition of SIRT6-deficient hMSCs after implantation. WT (1 × 106, left) and SIRT6-deficient (1 × 106, right) hMSCs (passage 6) pretransduced with luciferase were implanted into the muscles of mice. Luciferase activities were imaged and quantified 1 week after implantation. Data were presented as mean ± SEM, n = 4, ***P < 0.001.
Given that tissue stem cell exhaustion has been recently recognized as one of the hallmarks of aging10, and the Sirt6-deficient mice exhibit features of premature aging in mesodermal tissues20, we hypothesized that SIRT6 deficiency would result in accelerated attrition of hMSC pool. We thus differentiated the SIRT6−/− and WT hESCs into hMSCs. SIRT6−/− and WT hMSCs expressed typical hMSC markers such as CD73, CD90, and CD105 (Figure 1D), and were found negative for hMSC irrelevant antigens CD45, CD34 and CD43 (Supplementary information, Figure S1E)10,31. RT-qPCR and western blotting results confirmed the loss of both SIRT6 mRNA and protein in SIRT6−/− hMSC (Figure 1E and Supplementary information, Figure S1F). Immunofluorescence indicated a relatively diffused localization pattern of SIRT6 in the nuclei of WT hMSCs, which was absent in the SIRT6−/− hMSCs (Figure 1F).
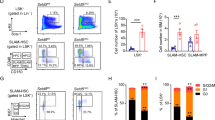
As the Sirt6-deficient mice exhibited bone abnormalities20, we wondered whether the differentiation potential was compromised in SIRT6−/− hMSCs. As expected, compared with WT hMSCs, SIRT6−/− hMSCs exhibited substantially impaired differentiation towards osteoblasts or chondrocytes (Supplementary information, Figure S1G). Next, we investigated the link between SIRT6 and hMSC aging. Although serial passaging of SIRT6−/− hMSCs led to several observations implicated in premature senescence such as early onset of senescence-associated (SA)-β-Gal activity (Figure 1G), upregulation of P16 and P21 proteins33 (Figure 1H), and progressive impairment in cell proliferation ability (Supplementary information, Figure S1H), SIRT6−/− hMSCs also exhibited features distinct from hMSCs derived from premature aging patients10. For instance, different from WS-specific hMSCs10, decreased transcripts from centromeric repetitive sequences34, and a mild but significant increase of G2/M population were observed in SIRT6−/− hMSCs (Supplementary information, Figure S1I and S1J). We ruled out the possibility that the premature aging phenotypes observed in SIRT6−/− hMSCs resulted from chromosomal instability as reported in mouse study20, as SIRT6−/− hMSCs exhibited normal karyotypes and minimal genomic mutational load determined by genome-wide copy number variation (CNV) analysis (Supplementary information, Figure S2A and S2B). Moreover, no significant increase in DNA damage response marker γH2AX was observed in SIRT6−/− hMSCs (Supplementary information, Figure S2C and S2D). Finally, we investigated whether SIRT6−/− hMSCs suffered from accelerated functional decay in an in vivo niche by transplanting WT and SIRT6−/− hMSCs into tibialis anterior (TA) muscle of the immunodeficient mice10. Compared with WT hMSCs, an accelerated in vivo decay of SIRT6−/− hMSCs was observed after engraftment (Figure 1I, Supplementary information, Figure S2E and S2F). The attrition of SIRT6−/− hMSCs could be alleviated by lentiviral vector-mediated reconstitution of exogenous SIRT6 (Supplementary information, Figure S2G). Altogether, these results support that SIRT6 has a key role in preventing hMSCs from premature attrition.
SIRT6−/− hMSCs are susceptible to oxidative stress
To unveil how SIRT6 deficiency results in the dysregulation of hMSC homeostasis, we evaluated the sensitivity of WT and SIRT6−/− hMSCs to various extrinsic stresses (Supplementary information, Figure S2H). PX-12, a potent inhibitor of thioredoxin and inducer of oxidative stress35,36, caused the strongest cytotoxicity in SIRT6−/− hMSCs, relative to WT hMSCs (Supplementary information, Figure S2H). Treatment with 50-100 μM of PX-12, concentrations known to effectively inhibit cellular thioredoxin37, led to significantly reduced viability in SIRT6−/− hMSCs by inducing apoptosis (Figure 2A, 2B and Supplementary information, Figure S2I). Similarly, SIRT6−/− hMSCs also exhibited increased susceptibility to cytotoxicity induced by paraquat, an inducer of endogenous ROS38,39 (Supplementary information, Figure S2J). We observed that SIRT6−/− hMSCs had higher basal levels of cellular ROS and DNA oxidation marker 8-oxo-7,8-dihydro-2′-deoxyguanosine (8-oxodG), relative to its WT counterparts (Figure 2C and 2D). Pretreatment of SIRT6−/− hMSCs with the antioxidant N-acetylcysteine (NAC) decreased the susceptibility to PX-12-induced cytotoxicity (Figure 2E), indicating that elevated cellular ROS level is in part responsible for the increased vulnerability of SIRT6−/− hMSCs to oxidative injury. Notably, reconstitution with WT SIRT6, but not the deacetylase-dead H133Y mutant24,26, not only diminished cellular ROS levels but also decreased the sensitivity of SIRT6−/− hMSCs to PX-12 treatment (Figure 2F–2H). In addition, reintroduction of WT SIRT6, but not the H133Y mutant into SIRT6−/− hMSCs repressed accelerated cellular senescence (Supplementary information, Figure S2K). Therefore, our data indicate that the SIRT6 deacetylase activity is crucial to counteract ROS and protect hMSCs from oxidative stress-associated functional decay.
Ablation of SIRT6 in hMSCs results in elevated ROS levels and increased vulnerability to oxidative injury. (A) WT and SIRT6-deficient hMSCs were treated with vehicle (DMSO) or 50 μM PX-12 for 24 h, and the apoptotic cells were determined by Annexin V-PI staining via FACS. (B) Statistical analysis of A. Apoptotic cell percentage in vehicle-treated WT hMSCs was normalized to 1. Data were presented as mean ± SEM, n = 3, NS, not significant, *P < 0.05. (C, D) Cellular reactive oxygen species (ROS) and 8-oxodG levels were determined by staining with H2DCFDA probe (C) and an anti-8-oxodG antibody (D), respectively, and measured by FACS. (E) SIRT6-deficient hMSCs were pretreated with H2O (control) or 1 mM NAC for 1 week, and then were treated with vehicle (DMSO) or 50 μM PX-12 for 24 h. Cellular apoptosis was measured by Annexin V-PI staining. Data were presented as mean ± SEM, n = 3, **P < 0.01. (F) Overexpression of WT SIRT6 (SIRT6 (WT)), not SIRT6 H133Y mutant (SIRT6 (HY)), in SIRT6-deficient hMSCs partially restored cellular ROS to normal levels. A luciferase (Control)-expressed vector was used as control. (G, H) SIRT6-deficient hMSCs were transduced with SIRT6 (WT), SIRT6 (HY), or Control vector, and then cells were treated with vehicle (DMSO) or 50 μM PX-12 for 24 h. Cell viability (G) and cellular apoptosis (H) were measured by MTS assay and Annexin V-PI staining, respectively. Data in G and H were presented as mean ± SEM, n = 6, *P < 0.05, **P < 0.01, ***P < 0.001.
SIRT6 positively regulates NRF2-mediated HO-1 expression in hMSCs
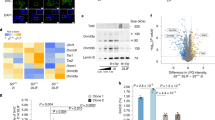
To uncover the molecular mechanism underlying how SIRT6 regulates redox homeostasis in hMSCs, we performed genome-wide RNA sequencing (RNA-seq) and H3K4me3 chromatin immunoprecipitation (ChIP) sequencing (ChIP-seq) analyses. We identified 119 upregulated genes and 246 downregulated genes in SIRT6−/− hMSCs relative to WT hMSCs (Figure 3A and Supplementary information, Figure S3A–S3C and Tables S1–S2). We analyzed biological pathways that were potentially altered due to SIRT6 deficiency and found that SIRT6−/− hMSCs had specific downregulation in genes associated with the GO terms such as “skeletal system development” (Figure 3B and Supplementary information, Figure S3D), which is in agreement with observed phenotypic defects in osteogenesis and chondrogenesis (Supplementary information, Figure S1G). Interestingly, the most significant GO term for downregulated genes was “response to oxygen levels” (P = 4.8E−8), which included a list of NRF2-regulated antioxidant genes16,40 (Figure 3C and 3D and Supplementary information, Figure S3D and S3E and Table S3). These genes were markedly downregulated in SIRT6−/− hMSCs (Figure 3D), which was associated with decreased levels of active chromatin mark H3K4me3 at their promoters (Figure 3E and Supplementary information, Figure S3F and S3G). In contrast to previous studies in mouse cells23,24,25,41, we did not observe global upregulation of HIF1α, c-MYC, c-JUN, and NF-κB target genes in SIRT6−/− hMSCs (Supplementary information, Figures S4A and S4B). In addition, SIRT6 deficiency-induced coordinated downregulation of NRF2 target genes appears to be specific to hMSCs, as we did not observe this in SIRT6−/− human vascular endothelial cells (hVECs) differentiated from SIRT6−/− hESCs (Supplementary information, Figure S5A–S5C).
SIRT6 is required for NRF2-depedent HO-1 expression in hMSCs. (A) Volcano plot showing significantly altered genes (q-value < 0.05, FC[SIRT6-deficient/WT] < 0.5 or FC [SIRT6-deficient/WT] > 2) between WT and SIRT6-deficient hMSCs. Representative NRF2 target genes were highlighted (indicated by arrows). FC, fold change. (B) Gene ontology (GO) analysis (biological process) of significantly downregulated genes in hMSCs upon SIRT6 depletion. (C) Venn diagram showing that early passage (EP, passage 6) and late passage (LP, passage 9) hMSCs shared 183 significantly downregulated genes in SIRT6-deficient hMSCs compared with WT hMSCs. NRF2 target genes shared in EP and LP were indicated. (D) RT-qPCR analysis of NRF2 target genes in WT and SIRT6-deficient hMSCs. Values were normalized against 18S rRNA. Data were presented as mean ± SEM, n = 3, *P < 0.05, **P < 0.01, ***P < 0.001. (E) Average profile of the H3K4me3 histone modification around the gene body regions of NRF2 target genes in SIRT6-deficient and WT hMSCs. TSS, transcription start site; TTS, transcription termination site. (F, G) RT-qPCR (F) and western blotting (G) analyses of HO-1 expression in WT and SIRT6-deficient hMSCs treated with 25 μM PX-12 for the indicated times. Relative mRNA and protein expressions were presented as fold induction. For RT-qPCR (F), values were normalized against 18S rRNA. Data were presented as mean ± SEM, n = 3, *P < 0.05, **P < 0.01. (H) Overexpression of SIRT6 (WT), not SIRT6 (HY), in SIRT6-deficient hMSCs partially restored HO-1 transcript. Values were normalized against 18S rRNA. Data were presented as mean ± SEM, n = 3, **P < 0.01, ***P < 0.001.
Among the NRF2-responsive genes downregulated in SIRT6−/− hMSCs, we chose to focus on HO-1 (also referred to as HMOX1), which encodes a potent antioxidative and cytoprotective factor42,43,44. We found that HO-1 was quickly induced by PX-12 treatment with mRNA level peaking at 4 h and protein level peaking at 8 h in WT hMSCs (Figure 3F and 3G), indicating that HO-1 is an early response gene induced by oxidative stress in hMSCs. In contrast, HO-1 expression level in SIRT6−/− hMSCs was insensitive to PX-12 treatment (Figure 3F and 3G), which was partially restored by overexpression of WT SIRT6, but not the H133Y mutant (Figure 3H). These data indicate that SIRT6 is required for the induction of HO-1 in response to oxidative stress.
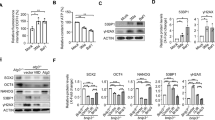
We next determined whether SIRT6 is directly involved in the NRF2-mediated transcription. We transfected SIRT6−/− and WT hMSCs with an ARE-driven luciferase reporter. Compared with WT, SIRT6 deficiency resulted in a significantly reduced luciferase activity in hMSCs (Figure 4A). Overexpression of WT SIRT6, but not its H133Y mutant, enhanced NRF2-mediated gene expression in primary hMSCs (Figure 4B and 4C), suggesting that SIRT6 positively regulated NRF2-mediated transcription in a deacetylase-dependent manner. There was no discernible difference in nuclear NRF2 levels between WT and SIRT6−/− hMSCs (Supplementary information, Figure S5D). ChIP-qPCR analysis also indicated that SIRT6 deficiency did not affect the amount of NRF2 associated with the classic ARE of endogenous HO-1 enhancer (Supplementary information, Figure S5E). These observations raised an intriguing possibility that SIRT6 directly coactivated NRF2-mediated transcription. To test this, we used the GAL4(DBD)-NRF2/(UAS)5-TATA-luciferase system. We found that the transactivation ability of NRF2 was positively regulated by SIRT6 in a deacetylase-dependent manner (Figure 4D and 4E). Using this system, we also identified that NRF2 was able to mediate recruitment of SIRT6 to the promoter DNA, the (UAS)5, GAL4-binding site by ChIP-qPCR (Figure 4F). Co-immunoprecipitation (Co-IP) and GST pull-down analyses showed that SIRT6 associated with NRF2 in a protein complex (Figure 4G, 4H and Supplementary information, Figure S5F and S5G).We next examined whether SIRT6 serves as a bridge between NRF2 and the basal transcriptional machinery. Co-IP experiment showed that SIRT6 formed a protein complex with RNA polymerase II (RNAP II) and TAF II-p135, two components of RNAP II complex (Figure 5A). ChIP-qPCR analysis further identified that both SIRT6 and RNAP II were recruited to the HO-1 promoter in WT hMSCs (Figure 5B and 5C). SIRT6 deficiency led to reduction of HO-1 promoter-bound RNAP II (Figure 5C). These data suggest that SIRT6 could help assemble NRF2-RNAP II transcription complex at the HO-1 promoter.
SIRT6 interacts with NRF2 and positively regulates NRF2-ARE pathway. (A) Transcriptional activity of NRF2 in WT and SIRT6-deficient hMSCs was measured by ARE-driven luciferase reporter assay. WT and SIRT6-deficient hMSCs were transfected with pcDNA3.1 (vector) or pcDNA3.1-NRF2 (NRF2), together with ARE-luciferase and Renilla plasmids. Data were presented as mean ± SEM, n = 3, *P < 0.05. (B) Plasmid expressing GFP, SIRT6 (WT), or SIRT6 (HY) was transfected into hMSCs, together with NRF2 or vector, and then NRF2 activity was measured using ARE-driven luciferase reporter. Data were presented as mean ± SEM, n = 3, NS, not significant, *P < 0.05. (C) Effect of SIRT6 overexpression on activation of NRF2 target genes in primary hMSCs. hMSCs were transduced with luciferase (control), SIRT6 (WT), SIRT6 (HY), or NRF2, and then the HO-1 and AKR1C1 transcripts were determined by RT-qPCR. Data were presented as mean ± SEM, n = 3, *P < 0.05, **P < 0.01, ***P < 0.001. (D) WT and SIRT6-deficient hMSCs were transfected with a (UAS)5-TATA-luciferase plasmid together with GAL4 or GAL4-NRF2, and then the NRF2 transactivity was measured. Data were presented as mean ± SEM, n = 3, NS, not significant, **P < 0.01. (E) Luciferase analysis of NRF2 transactivity in WT and SIRT6−/− hMSCs in the presence of overexpressed GFP, SIRT6 (WT), or SIRT6 (HY). Data were presented as mean ± SEM, n =3, *P < 0.05, **P < 0.01. (F) ChIP-qPCR analysis of (UAS)5-associated SIRT6 in hMSCs co-expressing (UAS)5-TATA-luciferase, GAL4-NRF2, and Flag-SIRT6 using an anti-Flag antibody. Data were presented as mean ± SEM, n = 3, **P < 0.01. (G) Co-immunoprecipitation (Co-IP) showing that SIRT6 and NRF2 formed a protein complex. Exogenous (upper and middle panels) and endogenous (lower panel) Co-IPs were performed with the indicated antibodies. (H) GST-NRF2 or GST protein expressed from E. coli was incubated with Flag-SIRT6 expressed from HEK293T cells. The GST pull-down assay indicated an in vitro interaction between NRF2 and SIRT6.
SIRT6 deacetylates H3K56 and is required for recruiting RNAP II to the HO-1 gene promoter. (A) Co-IP assay using protein extracts from HEK293T cells expressing Flag-SIRT6 indicated that SIRT6 formed a protein complex with RNAP II and TAF II-p135. (B) ChIP-qPCR performed in SIRT6−/− hMSCs transduced with Flag-SIRT6 or Flag-luciferase (control) indicated association of SIRT6 with HO-1 promoter and enhancers. Data were presented as mean ± SEM, n = 3, *P < 0.05, **P < 0.01. (C) ChIP-qPCR assay showing SIRT6-dependent recruitment of RNAP II at HO-1 promoter. Data were presented as mean ± SEM, n = 3, *P <0.05. (D) Western blotting analyses of H3K56Ac, H3K9Ac, and H3K4me3 in WT and SIRT6-deficient hMSCs. Histone 3 (H3) was used as the loading control. (E) Immunofluorescence (left) and statistical (right) analyses of H3K56Ac levels in WT and SIRT6-deficient hMSCs. Scale bar, 100 μm. (F) ChIP-qPCR analysis of the enrichment of H3K56Ac and H3K9Ac at HO-1 promoter in WT and SIRT6-deficient hMSCs. Data were presented as mean ± SEM, n = 3, NS, not significant, **P < 0.01. (G) ChIP-qPCR analysis of H3K56Ac at HO-1 promoter in WT or SIRT6-deficient hMSCs transduced with lentiviral vector encoding SIRT6 (WT), SIRT6 (HY), or luciferase (control). Data were presented as mean ± SEM, n = 3, **P < 0.01.
Deacetylation of H3K56 by SIRT6 accounts for recruitment of RNAP II to HO-1 promoter
In murine studies, SIRT6 was shown to act as a transcriptional repressor by deacetylating H3K9 at the gene promoters23,24,25. As SIRT6 deficiency did not result in upregulation of the H3K9Ac level, it is thus unlikely that H3K9Ac is the major substrate of SIRT6 in hMSCs (Figure 5D and Supplementary information, Figure S6A). In contrast, the level of H3K56Ac, another substrate of SIRT628,45, was dramatically increased in SIRT6−/− hMSCs (Figure 5D, 5E and Supplementary information, Figure S6A). In addition, ChIP-qPCR showed more enrichment of acetylated H3K56 at the promoters of NRF2-responsive genes including HO-1, AKR1C1, and PTGS2 in SIRT6−/− than in WT hMSCs (Figure 5F and Supplementary information, Figure S6B). We observed a reduced H3K56 acetylation level at HO-1 promoter when WT SIRT6 but not the H133Y mutant was reintroduced into SIRT6−/− hMSCs (Figure 5G and Supplementary information, Figure S6C). In addition, reintroduction of WT SIRT6 instead of the H133Y mutant increased the recruitment of RNAP II to the HO-1 promoter in SIRT6−/− hMSCs (Supplementary information, Figure S6D), suggesting a deacetylase activity-dependent recruitment of RNAP II by SIRT6. Next, we investigated whether deacetylation of H3K56 by SIRT6 at HO-1 promoter is causally linked to the recruitment of RNAP II. Ectopic expression of H3K56Q, an H3K56 acetylation-mimic mutant28, in WT hMSCs inhibited the recruitment of RNAP II to the HO-1 promoter (Supplementary information, Figure S6E). On the other hand, ectopic expression of H3K56R, an acetylation-defective mutant46, in SIRT6−/− hMSCs promoted occupancy of RNAP II at HO-1 promoter (Supplementary information, Figure S6E). Altogether, these data indicated that SIRT6 is responsible for the deacetylation of H3K56Ac at the HO-1 promoter in hMSCs, which is important for the recruitment of RNAP II transcriptional machinery.
SIRT6-NRF2-HO-1 pathway accounts for anti-oxidative response in hMSCs
As SIRT6−/− hMSCs exhibited increased basal ROS production and increased sensitivity to oxidative stress, next we tested whether the SIRT6-NRF2-HO-1 axis is linked to hMSCs' anti-oxidative response. We found that overexpression of HO-1 resulted in alleviation of ROS levels in SIRT6−/− hMSCs (Figure 6A). In addition, we observed a decrease in the susceptibility of SIRT6−/− hMSCs to PX-12-induced cytotoxicity upon HO-1 overexpression (Figure 6B and 6C). Finally, we investigated whether SIRT6-NRF2-HO-1 axis functions in the regulation of hMSC redox homeostasis in an in vivo context. To this end, SIRT6−/− hMSCs were transduced with a lentiviral HO-1 expression vector and then transplanted into the TA muscles of immunodeficient mice (Figure 6D and Supplementary information, Figure S6F). As expected, HO-1 overexpression repressed accelerated cell attrition of SIRT6-deficient hMSCs in an in vivo microenvironment (Figure 6D).
Compromised NRF2-HO-1 axis accounts for redox dysregulation in SIRT6-deficient hMSCs. (A) FACS analyses of ROS level in hMSCs transduced with lentiviral vector encoding luciferase (control) or HO-1. (B) Lactate dehydrogenase (LDH) detection in the indicated hMSCs transduced with lentiviral vector encoding luciferase (control) or HO-1 in the presence of 50 μM PX-12 treatment. Data were presented as mean ± SEM, n = 3, *P < 0.05. (C) FACS analyses of PX-12-induced cytotoxicity in hMSCs transduced with lentiviral vector encoding luciferase (control) or HO-1 (left panel). Cells were treated with vehicle (DMSO) or 50 μM PX-12 for 24 h. Statistical analysis of apoptotic cells (right panel) was presented as mean ± SEM. n = 6, **P < 0.01. (D) Measurement of luciferase activity in immunodeficient mice with IVIS. SIRT6-deficient hMSCs overexpressing GFP plus luciferase (control group, left) and SIRT6-deficient hMSCs overexpressing HO-1 plus luciferase (right) were implanted into the TA muscles of mice. Six days after implantation, mice were intraperitoneally injected with 20 mg/kg PX-12 for 24 h, and then luciferase activity was measured. Data were presented as mean ± SEM, n = 4, *P < 0.05. (E) A putative model for SIRT6-mediated redox regulation in hMSCs. In WT hMSCs, SIRT6 is a key regulator of the cellular redox homeostasis by co-activating NRF2 antioxidant pathway. SIRT6 associates with NRF2 and deacetylates H3K56 at the promoter of NRF2 target genes (i.e., HO-1), which is required for the recruitment of RNAP II complex and subsequent transactivation of NRF2. In SIRT6-deficient hMSCs, SIRT6 deficiency causes increased level of H3K56Ac and impaired recruitment of RNAP II complex to HO-1 promoter, resulting in decrease in HO-1 expression and compromised cellular redox homeostasis.
Disscusion
Throughout lifetime, tissue stem cells are constantly exposed to various stresses including ROS, toxins, and chemical or physical stressors, which may result in premature cellular aging or decreased cellular survival. Here, by employing the state-of-the-art genome-editing technology to specifically delete SIRT6 in human stem cells, we have presented several lines of evidence in support of a role of SIRT6 in safeguarding hMSCs from functional decay: (1) SIRT6 deficiency leads to an increase in ROS levels and vulnerability to oxidative insults; (2) SIRT6-depleted hMSCs show accelerated cellular senescence; (3) SIRT6 deficiency results in impairment of differentiation potential of hMSCs into bone and cartilage. These new findings establish a previously unappreciated connection between SIRT6, oxidative stress, and human stem cell exhaustion, and indicate that SIRT6-NRF2 module may be the key molecular hub to protect the mesodermal tissues from aging-associated degeneration.
Mechanistically, SIRT6 is identified as a positive regulator of NRF2-ARE antioxidant pathway in hMSCs (Figure 6E). We propose a model where SIRT6 plays dual roles in NRF2-mediated HO-1 transcription: (1) SIRT6 stabilizes the protein complex comprised of NRF2 and basal transcription machinery, potentially as an adaptor protein; (2) SIRT6 also exerts its histone deacetylase activity specifically at the promoter of HO-1. The two functions of SIRT6 are potentially linked as SIRT6-mediated deacetylation of H3K56 may be a prerequisite for stabilizing the NRF2-SIRT6-RNAP II complex at chromosomal levels.
To the best of our knowledge, this study provides the first evidence that SIRT6 can serve as a transcriptional coactivator. Recent murine studies showed that SIRT6 could transrepress the activities of HIF1α, c-JUN, and NF-κB via H3K9 deacetylation23,24,25. In hMSCs, however, we found that acetylated H3K9 is not a preferred substrate for SIRT6. Likewise, we did not observe global upregulation of the target genes of HIF1α, c-MYC, c-JUN, and NF-κB23,24,2541 in SIRT6-depleted hMSCs. Instead, deficiency of SIRT6 in hMSCs results in an increase in acetylated H3K56 levels, and SIRT6-mediated deacetylation of H3K56 is a key event facilitating NRF2-dependent gene expression. In addition, in line with a role of SIRT6 as a NRF2 coactivator, SIRT6 interacts with RNAP II complex and the depletion of SIRT6 in hMSCs diminishes the enrichment of RNAP II complex at the HO-1 promoter. Of note, a recent genome-wide study in hESCs revealed that SIRT6 co-localizes with RNAP II at gene promoter regions47, which together with our finding raises an interesting possibility that other SIRT6-mediated transcriptional activation events may also exist. In addition, it should be noted that SIRT6-mediated positive regulation of NRF2 activity appears to be specific to hMSCs. Indeed, we did not observe global downregulation of NRF2 target genes in SIRT6-deficient hVECs. In these contexts, more complex regulations such as cell-type-specific protein-protein interactions, deacetylation-independent activities of SIRT6, as well as potential compensations from other histone deacetylases may be involved.
The implication of SIRT6-regulated H3K56 acetylation in various cellular events has not been fully investigated. A very recent study indicated that human SIRT6 acts as a scaffold for recruiting SNF2H, a chromatin remodeler, and other DNA repair factors to DNA damage foci, an event dependent on SIRT6's deacetylase activity at histone H3K5628. It is likely that SIRT6-mediated low acetylation level of H3K56 at DNA damage site is important for an open local chromatin configuration necessary for DNA repair. Along this line of thinking, one could reason that transcriptional events are also aided by a permissible chromatin configuration, i.e., chromatin remodeling-dependent factor recruitment. Therefore, it is reasonable that SIRT6/NRF2-dependent HO-1 transcription may employ a similar H3K56 deacetylation-based mechanism to recruit RNAP II complex. How the increase in local H3K56Ac levels potentially regulates chromosomal configuration and recruitment of transcriptional machineries warrants further investigation.
The findings that SIRT6-NRF2-HO-1 axis acts as a regulator for stem cell redox homeostasis and SIRT6 functions as a transcriptional coactivator undoubtedly add a new layer of knowledge to SIRT6 biology. Safeguarding redox homeostasis by SIRT6 in hMSCs principally could contribute to mechanistic explanations for various SIRT6-related biological processes, including genomic instability, cellular senescence, cellular transformation, and metabolic dysregulation. Our study provides first evidence that SIRT6 is a key gatekeeper for human adult stem cell homeostasis, highlighting that SIRT6-NRF2 pathway may be a novel target for preventing aging-associated stem cell attrition, and hopefully, for treating aging-associated disorders in the future.
Materials and Methods
Cell culture
H9 hESCs (WiCell Research) were maintained on Mitomycin C-inactivated mouse embryonic fibroblast (MEF) or Matrigel31,48. hMSCs were cultured in DMEM (Hyclone) medium supplemented with 10% fetal bovine serum (FBS, AusGeneX), 0.1 mM non-essential amino acids (Gibco), 1% penicillin/streptomycin (Gibco), and 1 ng/ml bFGF (Joint Protein Central, JPC)31. Human primary MSCs were isolated from the teeth tissue of a 13-year-old male person and were cultured in hMSC culture medium. hVECs were cultured in EGM-2 (Lonza) medium containing VEGF165 (HumanZyme, 50 ng/ml) and FGF2 (JPC, 20 ng/ml).
Generation of SIRT6−/− hESCs
TALEN-mediated gene targeting was performed as previously described31. In brief, a donor plasmid was constructed by the combination of 1.0-2.0-kb homology arms and drug resistance cassettes (neo). H9 hESCs (5.0 × 106) were electroporated with a pair of TALEN vectors (Addgene, TAL2454 (Plasmid #36843) and TAL2455 (Plasmid #36844)) and the donor plasmid, and subsequently cultured on MEF feeder. G418 (100 μg/ml, Invitrogen) was then added to the medium to initiate positive selection 2-4 days after electroporation. After about 2 weeks' selection, G418-resistant clones were picked and transferred to a 96-well plate for further characterization and expansion. Gene-targeted clones were determined by genomic PCR with the primers listed in Supplementary information, Table S4 using PrimeSTAR DNA Polymerase (TAKARA). Long PCR cycling included a 1-min initial denaturation at 98 °C, 35 cycles of 10-s denaturation at 98 °C and a 0.5-3-min annealing and extension at 68 °C plus a final extension at 72 °C for 7 min.
Southern blotting
Genomic DNA was extracted following a described strategy48. 20 μg genomic DNA of each samples was digested with Hind III and Xho I (NEB) overnight and subjected to electrophoresis at 50 V on a 0.8% agarose gel (SeaKem Gold agarose, Lonza) for 3-4 h. The gel was subsequently incubated in 0.25 M HCl for 3-5 min followed by 2× 15-min incubation in denaturation buffer (0.5 M NaOH, 1.5 M NaCl) and 2× 15-min incubation in neutralization buffer (0.5 M Tris-HCl, pH 7.5; 1.5 M NaCl). The DNA was then blotted overnight onto a nylon membrane (GE Healthcare Life Sciences) by capillary transfer in 20× SSC buffer. The membrane was then ultraviolet crosslinked. The 5′ and 3′ probes were amplified from genomic DNA using a DIG-label kit (Roche) with the primers in Supplementary information, Table S4, following the manufacturer's protocol. The probes were labeled with DIG and Southern hybridization was performed following the standard protocol.
Excision of the neomycin-resistance cassette
To remove the neomycin-resistance cassette, SIRT6 gene-targeted hESCs were electroporated with pCAG-FLpo-2A-puro vector and then cultured on MEF feeder. Three days after transfection, puromycin (1 μg/ml; Invitrogen) was used to enrich puro-resistant cells. Puromycin was withdrawn after 48 h. Ten days later, the emerging colonies were picked and expanded in 96-well plates. Removal of the neomycin-resistance cassette was verified by PCR using PrimeSTAR DNA Polymerase (TAKARA) with the related primers (Supplementary information, Table S4).
hMSC generation and characterization
hMSCs were differentiated from hESCs based on a published protocol31. Briefly, embryoid bodies were left to differentiate in αMEM (Invitrogen) medium supplemented with 10% FBS (AusGeneX), 10 ng/ml bFGF (JPC), 5 ng/ml TGFβ (HumanZyme) and 1% penicillin/streptomycin (Gibco) until fibroblast-like cells appeared. The hMSCs were purified with different antibodies corresponding to hMSC-specific markers (CD73, CD90, and CD105) by FACS. Antibodies used for hMSC characterization were as follows: anti-CD105-APC (17-1057-42) antibody was purchased from eBioscience; anti-CD90-FITC (555595), anti-CD73-PE (550257), anti-CD34-PE (555822), anti-CD43-APC (580198), and anti-CD45-FITC (555482) antibodies were purchased from BD Biosciences. Anti-IgG-FITC (555748), anti-IgG-PE (555749), and anti-IgG-APC (555751) antibodies from BD Biosciences were used as isotype controls. The functionality of hMSC was further verified by differentiation towards cartilage, bone, and adipocytes31. The tri-lineage differentiation abilities of hMSC lines were evaluated by histochemical staining with von Kossa (osteogenesis), Alcian blue (chondrogenesis), and Oil red O (adiopogenesis) Kit (IHC World), respectively.
Directed differentiation of hESCs into hVECs
Differentiation was performed as previously described49, and the generated hVECs were characterized by the expression of hVEC-specific markers VE-cadherin, CD31, and VWF, as well as the activity of AC-LDL uptake.
Lentivirus preparation
The cDNAs of flag-SIRT6, flag-HO-1, flag-NRF2, Flag-luciferase, and flag-H3 were cloned into pLE4 lentiviral vector (a gift from Dr Tomoaki Hishida). pLE4-flag-SIRT6 (H133Y), pLE4-flag-H3K56Q, and pLE4-flag-H3K56R were generated using a fast mutagenesis kit (TransGen Biotech). Lentivirus particles were generated from HEK293T cells48, and used for transducing hMSCs in the presence of 4 μg/ml polybrene.
SA-β-GAL staining assay
Cells were stained using SA-β-GAL assay according to a previously described method48,50.
Clonal expansion assay
2 000 cells were seeded in each well of 12-well plates, cultured for 2 weeks, and stained with 0.2% crystal violet. Cell numbers were counted using light microscope in randomly selected fields. Each experiment was performed in triplicate.
Cell viability analysis
hMSCs (2 × 104) were seeded onto 0.1% gelatin-coated 96-well plates (Corning, Costar), and upon reaching 90% confluence cells were treated with various stressors for 24 h. Cell viability was measured using MTS approach according to the recommended protocol for CellTiter 96 AQueous One Solution Cell Proliferation Assay (Promega). Stressors used in this assay include DMSO (vehicle, Sigma), PX-12 (Santa Cruz Biotech), 4-nitroquinoline N-oxide (4NQO, Sigma), apoptosis activator 2 (AAT2, TOCRIS), ABT-737 (Santa Cruz Biotech), Mitomycin C (MMC, Sigma), and Stat3 inhibitor III (WP-1066, Santa Cruz), Paraquat (Sigma), NAC (Sigma).
Cell death analysis
Cell death was determined using Cytotox 96 non-radioactive cytotoxicity assay (Promega). Cells were treated with DMSO or 50 μM PX-12 for 24 h, and then the LDH level in culture medium was assayed. Each experiment was performed in biological triplicate.
Western blotting
Protein quantification was performed using a BCA Kit. Protein lysate was subjected to SDS-PAGE and subsequently electrotransferred to a polyvinylidene fluoride membrane (Millipore). Western blotting was performed as previously described8,48,51. The antibodies used are listed as follows (company, catalogue number, and dilution): anti-NRF2 (Abcam, ab62352, 1:1 000), anti-SIRT6 (Cell Signaling Technology, 12486, 1:1 000), anti-H3K9Ac (Millipore, 06-942, 1:1 000), anti-H3K56Ac (Abcam, ab76307, 1:5 000), anti-H3K4me3 (Active motif, 39159, 1:1 000), anti-P16 (Cell Signaling Technology, 4824, 1:200), anti-P21 (Santa Cruz, ZS-6246, 1:1 000), anti-γH2AX (Abcam, ab11175, 1:1 000), anti-HO-1 (Enzo, ADI-SPA-895-D, 1:1 000), anti-RNAP II (Covance, MMS-126R, 1:1 000), anti-β-actin (Santa Cruz, sc69879, 1:4 000), anti-lamin B1 (Abcam, ab16048, 1:1 000), anti-TAF II-p135 (Santa Cruz, sc136093, 1:1 000), anti-flag (Sigma, F1804, 1:4 000), and anti-HA (Santa Cruz, sc7392, 1:1 000).
Immunofluorescence
hESCs and hMSCs were fixed with formaldehyde (4% in PBS) for 15 min, permeabilized with Triton X-100 (0.4% in PBS) for 15 min, incubated with blocking buffer (10% donkey serum in PBS) for 30 min, and stained with primary antibody overnight at 4 °C. Then, the cells were treated with secondary antibodies for 1 h at room temperature. Hoechst 33342 (Invitrogen) was used to stain nuclear DNA. The antibodies used in immunofluorescence assay are as follows: anti-SIRT6 (CST, 12486, 1:100), anti-NANOG (Abcam, ab21624, 1:200), anti-SOX2 (Santa Cruz, sc-17320, 1:100), anti-OCT4 (Santa Cruz, sc-5279, 1:100), anti-Flag (Sigma, F1804, 1:1 000), anti-SMA (Sigma, A5228, 1:100), anti-β-tubulin III (TUJ1, Sigma, T2200, 1:100), anti-FOXA2 (CST, 8186S, 1:100), anti-VE-caderin (Cell Signaling Technology, 2158, 1:100), anti-CD31 (BD Pharmingen, 555445, 1:50), anti-VWF (Dako, A0082, 1:200), anti-Ac-LDL (Life Technology, l23380, 1:400).
Flow cytometry analysis
For cell cycle analysis, cells were pulsed with 10 μM EdU for 2 h, and collected using TrypLE Express (Invitrogen). Collected cells were stained with Alexa Fluor 647 dye azide and propidium iodide according to the manufacturer's instruction for Click-iT EdU Alexa Fluor 647 Flow Cytometry Assay Kit (Molecular Probes, c10419). Cell cycle profile was quantified by a FACS machine (BD LSRFortesa). For cell apoptosis analysis, cells were collected freshly and stained with Annexin V-EGFP and PI, and then apoptotic cells were quantified by FACS. For ROS measurement, cells were collected, loaded with 1 μM H2DCFDA for 30 min using ROS Detection Reagents (Molecular Probes, C6827), and quantified by FACS.
Co-IP assay
For exogenous Co-IP, cells were lysed in NP-40 lysis buffer (1% NP-40, 150 mM NaCl, 10% glycerol, 1 mM EGTA, 1 mM EDTA, 20 mM Tris, pH 7.5, and complete protease inhibitor cocktail (Roche)), and for endogenous Co-IP, cells were lysed in CHAPS buffer (120 mM NaCl, 0.3% CHAPS, 1 mM EDTA, 40 mM HEPES, pH 7.5, and complete protease inhibitor cocktail (Roche)). Cells were lysed on ice for 30 min, and then centrifuged at 12 000 rpm. at 4 °C for 20 min. For endogenous Co-IP, lysates (1 mg protein) were pre-cleared with 20 μl of Protein A/G-PLUS Agarose beads (Santa Cruz) for 2-4 h, and then the supernatants were collected by centrifugation at 3 000 rpm at 4 °C for 3 min. The supernatants mixed with the indicated antibodies and beads were rotated overnight at 4 °C. After being washed with NP-40 buffer or CHAPS buffer for three times, the immunocomplexes were eluted by boiling in 1× SDS-loading buffer for 10 min.
GST pull-down assay
Recombinant GST and GST-NRF2 proteins were expressed in BL21 cells (Genestar)52. Flag-SIRT6 protein was expressed in HEK293T. GST pull-down assay was performed as previously described53.
Luciferase reporter assay
For ARE-driven luciferase reporter system, hMSCs were co-transfected with 0.15 μg of ARE-driven luciferase, 0.15 μg pcDNA3.1-NRF2, 0.15 μg of cDNA expression vector, and 0.05 μg of renilla. For GAL4(DBD)-NRF2/(UAS)5-TATA luciferase reporter system, cells were co-transfected with 0.15 μg of (UAS)5-TATA luciferase, 0.05 μg of renilla, 0.15 μg of cDNA expression vector, and 0.15 μg of GAL4-fusion protein construct. At 48 h after transfection, cells were collected and relative luciferase activity was measured using Dual-Luciferase Reporter Assay System (Promega).
RNA-seq library construction
One million cells were applied to extract total RNA using the RNeasy Mini Kit (Qiagen) according to the manuals. After quantification of RNA by Fragment Analyzer (Advanced Analytical), 1.5 μg of total RNA was used to construct sequencing libraries by TruSeq RNA Sample Preparation Kit (Illumina) following the manufacturer's standard protocol.
Genomic library construction
Genomic DNA was extracted from 0.5-1 × 106 cells via DNeasy Blood & Tissue Kit (Qiagen). After being sheared into fragments ranging from 150 to 200 bp, the DNA was constructed into sequencing libraries using NEBNext DNA Library Prep Reagent Set for Illumina (NEB).
ChIP-seq
ChIP-seq was performed according to a previous protocol with some modifications54. Briefly, cells were crosslinked and lysed, and then the released chromatin was sheared into fragments. To obtain fragments of interest, samples were incubated with anti-H3K4me3 antibody (Abcam, ab8580) overnight. Inputs served as negative controls. After the DNA was de-crosslinked and extracted, the ChIP-seq libraries were generated using NEBNext DNA Library Prep Reagent Set for Illumina (NEB) according to manuals.
ChIP-seq data analysis
Clean reads were mapped to hg19 genome by BWA. Then, peaks were called by MACS255 by default parameters using input as control. Density of the promoter region was defined as the non-duplicate reads located in the regions between 2-kb upstream of the TSS (transcription start site) and 2-kb downstream of the TSS and then were normalized by the total non-duplicate reads that were mapped to the hg19 genome.
RNA-seq data analysis
Low-quality reads (N bases >10% of a read, >50% of the bases in a read with phred 33 < 5, or phred score of the first 5 bases < 20) were discarded. Then reads were mapped to the human reference genome hg19 (from UCSC) by Tophat56 and Cufflinks57. Transcript expression and differentially expressed genes were analyzed as previously reported58.
CNV analysis
Clean reads of genomic libraries were first mapped to hg19 genome by BWA59. Then, the reads in 0.5-M window along the genome were normalized by the total bases mapped to the reference and the number of non-N bases in 0.5-M genome window size.
RT-qPCR
Total RNA was extracted from cells according to the instruction of TRIzol Reagent (Invitrogen). 1-2 μg total RNA was used for cDNA synthesis with reverse transcription Master Mix (Promega). Quantitative real-time PCR was carried out using iTaq Universal SYBR Green Supermix (Bio-Rad) on a CFX384 Real-Time PCR system (Bio-Rad). All data were normalized by 18S rRNA transcript and calculated using the ΔΔCq method. All RT-qPCR primer pairs are listed in Supplementary information, Table S4.
ChIP-qPCR
ChIP-qPCR was performed according to a published protocol54. The antibodies used in ChIP-qPCR were as follows: anti-NRF2 (Abcam, ab62352, 4 μl), anti-RNAP II (Covance, MMS-126R, 4 μl), anti-H3K9Ac (Millipore, 06-942, 4 μl), anti-H3K56Ac (Abcam, ab76307, 2 μl), anti-flag (Sigma, F1804, 2.4 μl). The primers' sequences are listed in Supplementary information, Table S4.
Animal experiments
For mouse experiments, 6-8-week-old male NOD-SCID mice and immune-deficient CD-1 nude mice were used. Animal experiments performed in this study were approved by the Chinese Academy of Science Institutional Animal Care and Use Committee. For teratoma formation assay, NOD-SCID mice were injected s.c. with 3 × 106 WT and SIRT6-deficient hESCs in a Matrigel/mTeSR solution, respectively. Teratomas were collected when reaching a size of around 10 mm in diameter, and subjected to immunostaining. For hMSC transplantation assay, 106 WT and SIRT6-deficient hMSCs pretransduced with a lentiviral vector encoding luciferase were injected to TA muscle of CD-1 nude mice. Mice were imaged in vivo with IVIS spectrum imaging system (XENOGEN, Caliper) for luciferase activity. After imaging, TA muscle of mice was subjected to immunofluorescence analysis.
Statistical analysis
Results were presented as mean ± SEM. Two tailed Student's t-test was conducted using Graph-Pad Prism Software. P values < 0.05 were considered statistically significant (*).
Accession numbers
All of the RNA-seq and ChIP-seq data have been deposited in GEO under the accession number GSE64642. Whole-genome sequencing data for CNV analysis were deposited in SRA under the accession number SRP059859.
Author Contributions
HP, DG, and XL performed the majority of the experiments. HP performed the gene editing. DG and HP performed cell culture, differentiation, and cell functional analysis. LW, JQ, TY, JY, WZ, and GY performed cell culture and differentiation. FT, XL, and JL carried out the genomic and epigenomic analyses. HP and JZ performed the co-IP experiments. HP, DG, RR, WZ, YL, JY, YH, and HW performed animal experiments. JW, WZ, ZJ, ZM, and JL performed data analysis and wrote the manuscript. GHL, FT, JQ, HP, DG, and XL conceived this study and wrote the manuscript.
Competing Financial Interests
The authors declare no competing financial interests.
References
Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G . The hallmarks of aging. Cell 2013; 153:1194–1217.
Liu L, Cheung TH, Charville GW, et al. Chromatin modifications as determinants of muscle stem cell quiescence and chronological aging. Cell Rep 2013; 4:189–204.
Uccelli A, Moretta L, Pistoia V . Mesenchymal stem cells in health and disease. Nat Rev Immunol 2008; 8:726–736.
Lepperdinger G . Inflammation and mesenchymal stem cell aging. Curr Opin Immunol 2011; 23:518–524.
Liang R, Ghaffari S . Stem cells, redox signaling, and stem cell aging. Antioxid Redox Signal 2014; 20:1902–1916.
Stenderup K, Justesen J, Clausen C, Kassem M . Aging is associated with decreased maximal life span and accelerated senescence of bone marrow stromal cells. Bone 2003; 33:919–926.
Liu GH, Barkho BZ, Ruiz S, et al. Recapitulation of premature ageing with iPSCs from Hutchinson-Gilford progeria syndrome. Nature 2011; 472:221–225.
Liu GH, Qu J, Suzuki K, et al. Progressive degeneration of human neural stem cells caused by pathogenic LRRK2. Nature 2012; 491:603–607.
Yu KR, Kang KS . Aging-related genes in mesenchymal stem cells: a mini-review. Gerontology 2013; 59:557–563.
Zhang WQ, Li JY, Suzuki K, et al. A Werner syndrome stem cell model unveils heterochromatin alterations as a driver of human aging. Science 2015; 348:1160–1168.
Lavasani M, Robinson AR, Lu AP, et al. Muscle-derived stem/progenitor cell dysfunction limits healthspan and lifespan in a murine progeria model. Nat Commun 2012; 3:608.
Singh L, Brennan TA, Kim JH, et al. Brief report: long-term functional engraftment of mesenchymal progenitor cells in a mouse model of accelerated aging. Stem Cells 2013; 31:607–611.
Paul MK, Bisht B, Darmawan DO, et al. Dynamic changes in intracellular ROS levels regulate airway basal stem cell homeostasis through Nrf2-dependent Notch signaling. Cell stem cell 2014; 15:199–214.
Moore KA, Lemischka IR . Stem cells and their niches. Science 2006; 311:1880–1885.
Ray PD, Huang BW, Tsuji Y . Reactive oxygen species (ROS) homeostasis and redox regulation in cellular signaling. Cell Signal 2012; 24:981–990.
Gorrini C, Harris IS, Mak TW . Modulation of oxidative stress as an anticancer strategy. Nat Rev Drug Discov 2013; 12:931–947.
Bailey-Downs LC, Mitschelen M, Sosnowska D, et al. Liver-specific knockdown of IGF-1 decreases vascular oxidative stress resistance by impairing the Nrf2-dependent antioxidant response: a novel model of vascular aging. J Gerontol A Biol Sci Med Sci 2012; 67:313–329.
Valcarcel-Ares MN, Gautam T, Warrington JP, et al. Disruption of Nrf2 signaling impairs angiogenic capacity of endothelial cells: implications for microvascular aging. J Gerontol A Biol Sci Med Sci 2012; 67:821–829.
Mahlknecht U, Ho AD, Voelter-Mahlknecht S . Chromosomal organization and fluorescence in situ hybridization of the human Sirtuin 6 gene. Int J Oncol 2006; 28:447–456.
Mostoslavsky R, Chua KF, Lombard DB, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell 2006; 124:315–329.
Mao ZY, Hine C, Tian X, et al. SIRT6 promotes DNA repair under stress by activating PARP1. Science 2011; 332:1443–1446.
Van Meter M, Kashyap M, Rezazadeh S, et al. SIRT6 represses LINE1 retrotransposons by ribosylating KAP1 but this repression fails with stress and age. Nat Commun 2014; 5: 5011.
Kawahara TLA, Michishita E, Adler AS, et al. SIRT6 links histone H3 lysine 9 deacetylation to NF-kappa B-dependent gene expression and organismal life span. Cell 2009; 136:62–74.
Zhong L, D'Urso A, Toiber D, et al. The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1 alpha. Cell 2010; 140:280–293.
Sundaresan NR, Vasudevan P, Zhong L, et al. The sirtuin SIRT6 blocks IGF-Akt signaling and development of cardiac hypertrophy by targeting c-Jun. Nat Med 2012; 18:1643.
Michishita E, McCord RA, Berber E, et al. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature 2008; 452:492–496.
Yang B, Zwaans BM, Eckersdorff M, Lombard DB . The sirtuin SIRT6 deacetylates H3 K56Ac in vivo to promote genomic stability. Cell cycle 2009; 8:2662–2663.
Toiber D, Erdel F, Bouazoune K, et al. SIRT6 recruits SNF2H to DNA break sites, preventing genomic instability through chromatin remodeling. Mol Cell 2013; 51:454–468.
Michishita E, McCord RA, Boxer LD, et al. Cell cycle-dependent deacetylation of telomeric histone H3 lysine K56 by human SIRT6. Cell cycle 2009; 8:2664–2666.
Hockemeyer D, Wang HY, Kiani S, et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol 2011; 29:731–734.
Liu GH, Suzuki K, Li M, et al. Modelling Fanconi anemia pathogenesis and therapeutics using integration-free patient-derived iPSCs. Nat Commun 2014; 5:4330.
Pan H, Zhang W, Zhang W, Liu GH . Find and replace: editing human genome in pluripotent stem cells. Protein Cell 2011; 2:950–956.
Nagai K, Matsushita T, Matsuzaki T, et al. Depletion of SIRT6 causes cellular senescence, DNA damage, and telomere dysfunction in human chondrocytes. Osteoarthritis Cartilage 2015; 23:1412–1420.
Wang D, Zhou J, Liu X, et al. Methylation of SUV39H1 by SET7/9 results in heterochromatin relaxation and genome instability. Proc Natl Acad Sci USA 2013; 110:5516–5521.
You BR, Shin HR, Han BR, Park WH . PX-12 induces apoptosis in Calu-6 cells in an oxidative stress-dependent manner. Tumour Biol 2015; 36:2087–2095.
You BR, Shin HR, Park WH . PX-12 inhibits the growth of A549 lung cancer cells via G2/M phase arrest and ROS-dependent apoptosis. Int J Oncol 2014; 44:301–308.
Kunkel MW, Kirkpatrick DL, Johnson JI, Powis G . Cell line-directed screening assay for inhibitors of thioredoxin reductase signaling as potential anti-cancer drugs. Anticancer Drug Des 1997; 12:659–670.
Shibata M, Hakuno F, Yamanaka D, et al. Paraquat-induced oxidative stress represses phosphatidylinositol 3-kinase activities leading to impaired glucose uptake in 3T3-L1 adipocytes. J Biol Chem 2010; 285:20915–20925.
Castello PR, Drechsel DA, Patel M . Mitochondria are a major source of paraquat-induced reactive oxygen species production in the brain. J Biol Chem 2007; 282:14186–14193.
Malhotra D, Portales-Casamar E, Singh A, et al. Global mapping of binding sites for Nrf2 identifies novel targets in cell survival response through ChIP-Seq profiling and network analysis. Nucleic Acids Res 2010; 38:5718–5734.
Sebastian C, Zwaans BMM, Silberman DM, et al. The histone deacetylase SIRT6 is a tumor suppressor that controls cancer metabolism. Cell 2012; 151:1185–1199.
Suttner DM, Dennery PA . Reversal of HO-1 related cytoprotection with increased expression is due to reactive iron. FASEB J 1999; 13:1800–1809.
Parfenova H, Basuroy S, Bhattacharya S, et al. Glutamate induces oxidative stress and apoptosis in cerebral vascular endothelial cells: contributions of HO-1 and HO-2 to cytoprotection. Am J Physiol Cell Physiol 2006; 290:C1399–C1410.
Turkseven S, Kruger A, Mingone CJ, et al. Antioxidant mechanism of heme oxygenase-1 involves an increase in superoxide dismutase and catalase in experimental diabetes. Am J Physiol Heart Circ Physiol 2005; 289:H701–H707.
Schwer B, Schumacher B, Lombard DB, et al. Neural sirtuin 6 (Sirt6) ablation attenuates somatic growth and causes obesity. Proc Natl Acad Sci USA 2010; 107:21790–21794.
Li Q, Zhou H, Wurtele H, et al. Acetylation of histone H3 lysine 56 regulates replication-coupled nucleosome assembly. Cell 2008; 134:244–255.
Ram O, Goren A, Amit I, et al. Combinatorial patterning of chromatin regulators uncovered by genome-wide location analysis in human cells. Cell 2011; 147:1628–1639.
Liu GH, Suzuki K, Qu J, et al. Targeted gene correction of laminopathy-associated LMNA mutations in patient-specific iPSCs. Cell Stem Cell 2011; 8:688–694.
Patsch C, Challet-Meylan L, Thoma EC, et al. Generation of vascular endothelial and smooth muscle cells from human pluripotent stem cells. Nat Cell Biol 2015; 17:994–U294.
Debacq-Chainiaux F, Erusalimsky JD, Campisi J, Toussaint O . Protocols to detect senescence-associated beta-galactosidase (SA-betagal) activity, a biomarker of senescent cells in culture and in vivo. Nat Protoc 2009; 4:1798–1806.
Kawata K, Kobayashi Y, Souda K, et al. Enhanced hepatic Nrf2 activation after ursodeoxycholic acid treatment in patients with primary biliary cirrhosis. Antioxid Redox Signal 2010; 13:259–268.
Cullinan SB, Gordan JD, Jin JO, Harper JW, Diehl JA . The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: Oxidative stress sensing by a Cul3-Keap1 ligase. Mol Cell Biol 2004; 24:8477–8486.
Shi XB, Hong T, Walter KL, et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature 2006; 442:96–99.
Dahl JA, Collas P . A rapid micro chromatin immunoprecipitation assay (mu ChIP). Nat Protoc 2008; 3:1032–1045.
Zhang Y, Liu T, Meyer CA, et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol 2008; 9:R137.
Trapnell C, Pachter L, Salzberg SL . TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 2009; 25:1105–1111.
Roberts A, Pimentel H, Trapnell C, Pachter L . Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics 2011; 27:2325–2329.
Trapnell C, Roberts A, Goff L, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 2012; 7:562–578.
Li H, Durbin R . Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010; 26:589–595.
Acknowledgements
We are grateful to Yang Zhao for administrative assistance. This work was supported by the National Basic Research Program of China (973 Program; 2015CB964800, 2014CB910503, 2014CB964600 and 2012CB966704), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA01020312), the National High Technology Research and Development Program of China (2015AA020307), the National Natural Science Foundation of China (81330008, 31222039, 31201111, 81371342, 81300261, 81300677, 81271266, 81471414, 81422017, 81401159, 81420108017, 81525010, and 1322039), Beijing Natural Science Foundation (7141005 and 5142016), Program of Beijing Municipal Science and Technology Commission (Z151100003915072), Key Research Program of the Chinese Academy of Sciences (KJZDEW-TZ-L05), the Thousand Young Talents Program of China, National Laboratory of Biomacromolecules (012kf02; 2013kf05; 2013kf11; 2014kf02; 2015kf10), and State Key Laboratory of Drug Research (SIMM1302KF-17), China Postdoctoral Science Foundation Grant (2013M530751), and China Postdoctoral Science Foundation Funded project (2015T80142). WZ was supported by NIH grants K99/R00 CA158055, the V Scholar award, American Cancer Society seed grant, Breast Cancer Research Award and Oberley Award (National Cancer Institute Award P30CA086862) from Holden Comprehensive Cancer Center at the University of Iowa, and startup funds from the Department of Pathology, University of Iowa.
Author information
Authors and Affiliations
Corresponding authors
Additional information
( Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary information
Supplementary information, Figure S1
Characterization of SIRT6-deficient hESCs and differentiation of hESCs to hMSCs. (PDF 239 kb)
Supplementary information, Figure S2
Characterization of SIRT6-deficient hMSCs. (PDF 237 kb)
Supplementary information, Figure S3
Gene expression analyses of WT and SIRT6-deficient hMSCs. (PDF 319 kb)
Supplementary information, Figure S4
Gene expression and epigenome analyses of WT and SIRT6-deficient hMSCs. (PDF 198 kb)
Supplementary information, Figure S5
SIRT6 regulates recruitment of RNAP II to HO-1 promoter in a deacetylase-dependent manner. (PDF 213 kb)
Supplementary information, Figure S6
HO-1 overexpression rescues premature attrition of SIRT6-deficient hMSCs in vivo. (PDF 160 kb)
Supplementary information, Table S1
Up- and downregulated genes in SIRT6−/− hMSCs relative to WT hMSCs (early passage). (XLSX 30 kb)
Supplementary information, Table S2
Up- and downregulated genes in SIRT6−/− hMSCs relative to WT hMSCs (late passage). (XLSX 40 kb)
Supplementary information, Table S3
GO terms for downregulated genes in SIRT6−/− hMSCs. (XLSX 50 kb)
Supplementary information, Table S4
Sequences of primers and probes used in the study. (XLSX 17 kb)
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 Unported License. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Pan, H., Guan, D., Liu, X. et al. SIRT6 safeguards human mesenchymal stem cells from oxidative stress by coactivating NRF2. Cell Res 26, 190–205 (2016). https://doi.org/10.1038/cr.2016.4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cr.2016.4
Keywords
This article is cited by
-
Quercetin stimulates trophoblast fusion via the mitochondrial function
Scientific Reports (2024)
-
CRL2APPBP2-mediated TSPYL2 degradation counteracts human mesenchymal stem cell senescence
Science China Life Sciences (2024)
-
SIRT3/6: an amazing challenge and opportunity in the fight against fibrosis and aging
Cellular and Molecular Life Sciences (2024)
-
Microglia Sirt6 modulates the transcriptional activity of NRF2 to ameliorate high-fat diet-induced obesity
Molecular Medicine (2023)
-
Deacetylation of FOXP1 by HDAC7 potentiates self-renewal of mesenchymal stem cells
Stem Cell Research & Therapy (2023)