Abstract
YAP and TAZ are transcriptional co-activators and function as the major effectors of the Hippo tumor suppressor pathway, which controls cell growth, tissue homeostasis, and organ size. Here we show that YAP/TAZ play an essential role in amino acid-induced mTORC1 activation, particularly under nutrient-limiting conditions. Mechanistically, YAP/TAZ act via the TEAD transcription factors to induce expression of the high-affinity leucine transporter LAT1, which is a heterodimeric complex of SLC7A5 and SLC3A2. Deletion of YAP/TAZ abolishes expression of LAT1 and reduces leucine uptake. Re-expression of SLC7A5 in YAP/TAZ knockout cells restores leucine uptake and mTORC1 activation. Moreover, SLC7A5 knockout cells phenocopies YAP/TAZ knockout cells which exhibit defective mTORC1 activation in response to amino acids. We further demonstrate that YAP/TAZ act through SLC7A5 to provide cells with a competitive growth advantage. Our study provides molecular insight into the mechanism of YAP/TAZ target genes in cell growth regulation.
Similar content being viewed by others
Introduction
The Hippo pathway controls cell growth, tissue homeostasis, and organ size through LATS1/2-mediated phosphorylation and inhibition of the transcriptional co-activators yes-associated protein (YAP) and transcriptional coactivator with a PDZ-binding domain (TAZ)1,2,3,4,5. The LATS-dependent phosphorylation leads to cytoplasmic retention and degradation of YAP/TAZ. In the nucleus, YAP/TAZ complex with transcription factors, most prominently the TEA domain family members (TEADs), to induce gene activation6,7,8. Hyperactivation of YAP/TAZ leads to overgrowth and cancer9,10,11,12,13,14,15,16,17,18, but how YAP/TAZ confer cellular growth advantages is currently not well-understood2.
The mechanistic target of rapamycin complex 1 (mTORC1) is a central regulator of cell growth19,20,21, and its activation status is tightly regulated. A wide range of signals, including growth factors, nutrients, and stress conditions, have been shown to regulate mTORC1 activity. mTORC1 hyperactivation is frequently observed in cancer and inhibitors of mTORC1 are used in the clinic for treating certain cancers19,20,21. Amino acids (AAs) are among the most potent regulators of mTORC1. Once inside the cell, AAs act primarily through the Rag GTPases to recruit mTORC1 to the lysosome, where it is activated by Rheb GTPases, which integrate upstream growth factor signals to mTORC119,20,21.
In this study, we show that YAP/TAZ potentiate mTORC1 activity by increasing expression of the high-affinity L-type amino acid transporter (LAT1, also known as CD98), which is a heterodimer of SLC7A5 and SLC3A2. YAP/TAZ and TEAD directly induce transcription of SLC7A5, which rescues SLC3A2 protein expression by dimer formation, to increase LAT1 expression and amino acid uptake. The enhanced capacity of AA uptake provides a competitive growth advantage for YAP/TAZ cells, thereby linking YAP/TAZ target genes to mTORC1 activity and cell growth.
Results
YAP and TAZ modulate amino acid-induced mTORC1 activation
In mammalian cells, AAs are potent regulators of mTORC119,20,21, and different AAs activate mTORC1 via different mechanisms. For example, leucine (Leu) rapidly stimulates mTORC1 via the Rag GTPases, while glutamine (Gln) slowly activates mTORC1 via Arf1 GTPase21,22,23,24,25. Gln also facilitates Leu uptake to stimulate mTORC126. AA starvation, even in the presence of fresh serum, shuts down mTOR activity23,27,28. Therefore, because AAs are such a potent regulator of mTOR, we set out to delineate the mechanisms of this regulation. We examined mTORC1 activation by Gln and Leu in AA-starved cells in the presence of dialyzed serum (10%). Consistent with previous observations, short-term Gln treatment did not significantly induce mTORC1 activation (Supplementary information, Figure S1A), although pre-incubation with Gln strongly potentiated mTORC1 activation by Leu even when Leu concentration was ten times lower than that of normal DMEM medium (Figure 1A and Supplementary information, Figure S1A). These are the conditions for Gln/Leu treatment that were used throughout the study.
YAP and TAZ modulate AA-induced mTORC1 activation. (A) Glutamine (Gln) potentiates mTORC1 activation by leucine (Leu). HEK293A cells were AA starved for 6 h, then Gln stimulated for 30 min followed by 10 min of Leu stimulation. Concentrations of 0.01×, 0.1×, and 1× denote 1%, 10%, and 100%, respectively, of the leucine concentration in normal DMEM medium. mTORC1 activity is determined by western blotting for the phosphorylation of S6K, S6, and 4E-BP1. L.e. and s.e. denote long exposure and short exposure, respectively. (B) YAP/TAZ are required for mTORC1 activation by AAs. Wild-type and YAP/ TAZ double knockout (Y/T dbKO) 293A cells were co-cultured, AA starved, and stimulated with Gln and Leu as indicated. Cells were AA starved (top row), or stimulated by 1× Gln (second row), or 1× Gln and 0.1× Leu (Gln/Leu, third row), or treated with rapamycin and Gln/Leu (bottom row). Some Y/T dbKO cells, as indicated by the lack of YAP/TAZ labeling, are marked with white arrows. Note, that upon Gln/Leu stimulation only wild-type (positive YAP/TAZ labeling) cells show positive pS6 signal, a marker of active mTORC1. See Supplementary information, Figure S1D for full size image. Scale bar, 20 μm. (C) AA-induced S6 phosphorylation depends on YAP/TAZ. The fraction of pS6-positive cells per image of each genotype was quantified in experiments similar to B. n >75 cells of each genotype, per treatment. #1 and #2 denotes two independent Y/T dbKO clones. (D) YAP and TAZ dictates glutamine-potentiated leucine stimulation of mTORC1. Western blots of cell lysates from 293A WT cells and Y/T dbKO cells. Cells were AA starved for 6 h and then stimulated with 1× Gln followed by 0.1× Leu, or only 0.1× Leu. The western blot was probed to assess mTORC1 activity. Western blots were also performed for expression of YAP, TAZ, and the LAT1 high-affinity leucine transporter (comprised of SLC7A5 and SLC3A2). Cyr61 is a known target gene of YAP/TAZ, whereas vinculin (Vinc) serves a loading control.
We wished to examine whether YAP/TAZ modulate mTORC1 activation in response to AA. We generated YAP/TAZ double knockout (Y/T dbKO) 293a cells using CRISPR genomic editing technology (see Supplementary information, Figure S1B). YAP/TAZ knockout was confirmed by western blot (Supplementary information, Figure S1B-S1G). mTORC1 activity is sensitive to the cell culture conditions, including levels of nutrients and growth factors in the growth medium. To carefully compare mTORC1 activity between wild-type (WT) and Y/T dbKO cells, we performed co-culture to ensure identical culture conditions for both WT and dbKO cells. We initially utilized phosphorylated S6 (pS6) as a readout for mTORC1 activation and the lack of YAP/TAZ labeling as a marker for Y/T dbKO cells. We found that Gln/Leu stimulation strongly induced S6 phosphorylation in WT cells (Figure 1B-1D and Supplementary information, Figure S1D-S1G), but, surprisingly, failed to induce S6 phosphorylation in Y/T dbKO cells (Figure 1B-1D). To validate that Gln/Leu stimulation of pS6 was dependent on mTORC1, we pretreated cells with the mTORC1 inhibitor rapamycin before stimulating with Gln/Leu and found that rapamycin completely blocked phosphorylation of S6 (Figure 1B and 1C). To further confirm that mTORC1 activation was indeed impaired, we treated and prepared WT and Y/T dbKO cells separately and blotted for p4E-BP1 and pS6K, two direct substrates of mTORC123,28,29,30. Indeed, mTORC1 activation in Y/T dbKO cells was severely impaired upon Gln/Leu stimulation (Figure 1D). The above observations show that YAP/TAZ have a critical role in mTORC1 activation by AAs.
Glutamine potentiates leucine uptake to activate mTORC1. LAT1, which is the major high-affinity Leu transporter, has previously been implicated in Gln/Leu-mediated mTORC1 activation26,31,32. LAT1 is a heterodimer consisting of SLC7A5 and SLC3A2 and functions as an AA exchanger, transporting Leu (and other large neutral AA) into the cell while transporting Gln out of the cell26,32,33,34. We found that Y/T dbKO cells lost protein expression of both SLC7A5 and SLC3A2 (Figure 1D). Interestingly, the loss of SLC7A5 expression was consistently more severe in single YAP KO cells than in TAZ KO cells (Supplementary information, Figure S1C). As Y/T dbKO cells had the most severe phenotype (Supplementary information, Figure S1C), and YAP and TAZ have overlapping functions1, we focused on Y/T dbKO cells for the remainder of the study. These results suggest that the lack of LAT1 in Y/T dbKO cells may contribute to the defective mTORC1 activation in response to Gln/Leu stimulation.
YAP and TAZ regulate leucine uptake via SLC7A5
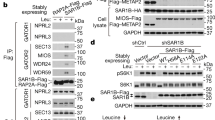
The LAT1 heterodimer is formed by a disulfide bond between SLC7A5 and SLC3A233. We examined for LAT1 heterodimer expression in WT and Y/T dbKO cells. The expression of LAT1 was strikingly lower in Y/T dbKO cells than in WT cells (Figure 2A). Treatment with the reducing agent β-mercaptoethanol abolished the high molecular weight dimer and produced the expected monomers of both SLC7A5 and SLC3A2. Re-expressing YAP in the Y/T dbKO cells restored expression of both SLC7A5 and SLC3A2, further confirming the key role of YAP/TAZ in regulating LAT1 expression. YAP/TAZ are negatively regulated by the LATS1/2 kinases1. We recently generated LATS1/2 dbKO 293a cells35,36, which results in nucleus-localized and hyperactivated YAP/TAZ (Supplementary information, Figure S1G). Deleting LATS1/2 resulted in a modest, but reproducible, increase in SLC7A5 and SLC3A2 (Figure 2A). Collectively, the above data establish an essential role for the Hippo pathway and YAP/TAZ in regulating the expression of SLC7A5 and SLC3A2.
YAP and TAZ regulates leucine uptake via SLC7A5. (A) YAP and TAZ are essential for LAT1 expression. LAT1 is a hetero dimeric transporter complex composed of SLC7A5 and SLC3A2, which are linked via a disulfide bond. Cell lysates from different cell lines as depicted in the box to the right were prepared in either non-reducing buffer (no β-mercaptoethanol) or reducing buffer (containing β-mercaptoethanol). Note that both the SLC7A5 antibodies and the SLC3A2 antibodies detect the high molecular heterodimeric complex under non-reducing condition (left part). Under the reducing condition (right half), both SLC7A5 and SLC3A2 run at the expected molecular weights of individual monomers. Ectopic expression of YAP (sample #a) rescues protein expression of both SLC3A2 and SLC7A5 in Y/T dbKO cells. (B) YAP/TAZ knockout decreases leucine uptake. H3-leucine uptake assay was carried out in cell lines (denoted in the right panel of Figure 2A) and normalized to WT cells. Error bars are means ± SEM of triplicates from a representative experiment. (C) LATS1/2 regulate leucine uptake via SLC7A5. Stable cell lines (right panel in Figure 2A) were generated and uptake of H3-leucine is displayed as relative to WT cells. Error bars are means ± SEM of triplicates from a representative experiment. (D) YAP/TAZ do not affect the plasma membrane localization of SLC7A5. WT and Y/T db KO cells that both stably express V5 epitope tagged SLC7A5 were co-cultured. YAP/TAZ WT cells are marked by positive YAP signal and dbKO cells (no YAP signal) are highlighted with white arrows. Ectopic SLC7A5 is localized to the plasma membrane in both WT and YAP/TAZ dbKO cells. Scale bar, 10 μm. (E) Reintroduction of SLC7A5 rescues the expression of SLC3A2 in YAP/TAZ knockout cells. Western blots of cell lysates from different cell lines as depicted. Re-expression of V5-tagged SLC7A5 or the epithelial specific SLC7A7, which likewise complexes with SLC3A2, but is not endogenously expressed in 293A cells, rescues the protein expression of SLC3A2. See also Supplementary information, Figure S3B. (F) Reintroduction of SLC7A5 restores leucine uptake in YAP/TAZ knockout cells. H3-leucine uptake assay was carried out in the indicated cell lines. #1 and #2 denotes two independent Y/T dbKO clones. SLC7A5 was re-expressed in Y/T dbKO clone #1. Error bars are means ± SEM of triplicates from a representative experiment.
The dramatic reduction of LAT1 expression indicates that Y/T dbKO cells may have compromised Leu transport. We performed Leu import assays with H3-labeled Leu. Indeed, we found that Y/T dbKO cells showed dramatically decreased Leu uptake (Figure 2B). As expected, re-expression of YAP in Y/T dbKO cells restored Leu uptake. The LATS1/2 dbKO cells also showed an increase in Leu uptake (Figure 2C). Knockdown of SLC7A5 decreased Leu uptake in LATS1/2 dbKO cells, suggesting that high LAT1 activity was responsible for the elevated Leu uptake in these cells (Figure 2C).
The dimeric LAT1 transporter complex functions at the plasma membrane (PM)26,33,34, as well as at the lysosome37. To exclude the possibility that LAT1 mislocalization may contribute to the reduced Leu uptake in Y/T dbKO cells, we examined the LAT1 cellular localization. We found that the ectopically expressed SLC7A5 was predominantly localized to the PM in both WT and Y/T dbKO cells (Figure 2D and Supplementary information, Figure S2) in accordance with its role in uptake of extracellular Leu.
SLC7A5 needs to be shuttled to the PM to be stable, and this is dependent on SLC3A2 expression33. When SLC7A5 was re-expressed in Y/T dbKO cells, it also dramatically increased SLC3A2 protein levels (Figure 2E), whereas re-expression of SLC3A2 did not increase SLC7A5 expression (Supplementary information, Figure S3A). These results suggest that YAP/TAZ regulate expression of SLC7A5, while the effect on SLC3A2 is possibly due to an indirect effect on protein stability of SLC3A2 by dimer formation with SLC7A5. Consistent with this hypothesis, exogenous expression of an epithelial-specific transporter subunit SLC7A738,39, which also complexes with SLC3A2 (Supplementary information, Figure S3B) but is not endogenously expressed in 293a cells, similarly increased SLC3A2 protein levels in Y/T dbKO cells (Figure 2E). To determine whether the reduced Leu uptake in Y/T dbKO cells was due to the decreased expression of SLC7A5, we performed Leu uptake assays in the SLC7A5 re-expressing Y/T dbKO cells. Our data showed that Leu uptake was rescued in Y/T dbKO cells when SLC7A5 was re-expressed (Figure 2F). Together, our study indicates that YAP/TAZ regulate SLC7A5 expression, and hence LAT1 activity, to promote Leu uptake.
To assess the function of SLC7A5 in WT cells, we generated SLC7A5 KO cells using CRISPR and investigated mTORC1 activation. Interestingly, following Gln/Leu stimulation, with regard to mTORC1 activation, the SLC7A5 KO cells phenocopies in great part Y/T dbKO cells (compare Figures 1D with 3A, and 1C with 3E, respectively). SLC7A5 KO cells displayed compromised phosphorylation of S6K, S6, and 4EBP1, especially under low Leu concentration (Figure 3A). Noteworthy, the effect of SLC7A5 knockout on mTORC1 activation was specific to AA stimulation, as the SLC7A5 KO cells had a normal response to insulin-induced mTORC1 activation (Figure 3B). Moreover, SLC7A5 KO also dramatically reduced SLC3A2 protein levels (Figure 3B). Similar results were obtained by pS6 labeling of WT and SLC7A5 KO cells (Figure 3C and 3E). These results are consistent with the notion that LAT1 is the major, high-affinity Leu transporter, while other low-affinity Leu transporters and other import mechanisms should still function in the SLC7A5 KO cells39,40. In addition, we measured Leu uptake in the SLC7A5 KO cells and confirmed that these cells are defective in Leu uptake (Figure 3F), confirming a vital role for LAT1 in cellular Leu uptake. The Rag GTPases are also important for mTORC1 activation by AAs22,41. As expected, RagA/B dbKO cells exhibited defective mTORC1 activation in response to Gln/Leu stimulation (Supplementary information, Figure S3C), consistent with the notion that SLC7A5 acts through the Rags to activate mTORC1.
SLC7A5 is required for AA signaling to mTORC1. (A) SLC7A5 is involved in Gln/Leu-induced mTORC1 activation. Wild-type and SLC7A5 knockout (KO) 293A cells were AA starved for 6 h and Gln stimulated for 30 min followed by 10 min of Leu stimulation at different concentrations. SLC7A5-deficient cells, which are confirmed by the lack of SLC7A5 expression, are impaired in mTORC1 activation at low Leu concentrations. Similar results were observed in an additional cell line independently generated (data not shown). (B) SLC7A5 does not affect insulin-induced mTORC1 activation. Experiments were similar to A except that cells were serum starved and stimulated with 100 nM insulin for the times illustrated, with or without 1 h pretreatment of mTOR inhibitors Torin (Tor) or Rapamycin (Rapa). Note also the lack of SLC3A2 expression in SLC7A5 KO cells. Similar results were observed in an additional cell line independently generated with a separate sgRNA targeting SLC7A5 (data not shown). (C) Representative confocal image of Gln/Leu-stimulated 293A WT cells, labeled for pS6 and stained for DAPI. Scale bar, 20 μm. (D) Confocal image of Gln/Leu-stimulated SLC7A5 KO 293A cells, labeled for pS6 and stained for DAPI. Samples and images in C and D were processed in parallel and acquired with the same microscope settings. Scale bar, 20 μm. (E) Leucine-induced mTORC1 activation is diminished in SLC7A5 KO cells. Quantification of confocal images as in C and D, and plotted as fraction of pS6-positive cells per image. The treatments were AA starved, AA starved then 1× Gln stimulated, or Gln/Leu stimulated, or pretreated with rapamycin for 1 h and then stimulated with Gln/Leu. (F) SLC7A5 is essential for efficient leucine uptake. H3-leucine uptake assay was carried out for three independently generated SLC7A5 KO clones and normalized to WT. Error bars are means ± SEM of triplicates from a representative experiment.
SLC7A5 is a direct YAP/TAZ-TEAD target gene to promote leucine uptake
As Y/T dbKO cells show a dramatic reduction in SLC7A5 protein levels (Figures 1D, 2A and 2E), we sought to determine whether this was due to regulation at the transcriptional level. Quantitative RT-PCR showed that Y/T dbKO cells expressed dramatically lower levels of SLC7A5 mRNA, whereas LATS1/2 dbKO cells had a slight increase in SLC7A5 mRNA expression (Figure 4A and 4B). YAP and TAZ single KO cells also showed a decrease in SLC7A5 mRNA expression (Supplementary information, Figure S3D). SLC7A5 mRNA levels were consistently lower in the YAP KO cells than in the TAZ KO cells, although this disparity was not conserved for other known YAP/TAZ target genes (Supplementary information, Figure S3D), as for these genes TAZ appeared more important. This observation suggests that YAP and TAZ may not have completely overlapping transcriptional profiles, highlighting the potential distinct functional importance of YAP and TAZ in downstream target gene expression. As YAP and TAZ are transcriptional coactivators, and the TEAD transcription factors are the major mediators of YAP/TAZ activity6,7,8, we therefore examined the upstream promoter region of SLC7A5 and identified a range of putative TEAD recognition motifs (Figure 4C). We cloned the SLC7A5 promoter regions into luciferase reporters. The reporters were co-transfected with or without YAP into the Y/T dbKO cells. We found that YAP potently activated the long reporter (with putative TEAD-binding sites), but not the short reporter (devoid of putative TEAD-binding sites), though both had similar basal activity (Figure 4D). Therefore, YAP stimulates SLC7A5 promoter activity.
SLC7A5 is a direct YAP/TAZ TEAD target gene to promote leucine uptake. (A) SLC7A5 expression is dependent on YAP/TAZ. qPCR of SLC7A5 mRNA levels in 293A WT cells compared to Y/T dbKO cells. (B) LATS1/2 dbKO increases SLC7A5 expression. qPCR of SLC7A5 mRNA levels from 293A WT cells compared to LATS1/2 dbKO cells. (C) The promoter of SLC7A5 contains several putative TEAD recognition motifs. Diagram depicting the upstream 2 kB of the SLC7A5 gene. TSS: transcription start site. ATG: start codon. Potential TEAD recognition motifs (CATTCC) are depicted with arrow heads. (D) YAP drives SLC7A5 promoter activity. The short (−1 075 to 0) or long (−2 000 to 0) fragment of SLC7A5 promoter region was fused to produce the luciferase reporters. These reporters were transfected into Y/T dbKO 293A cells in combination with YAP or vector control. Luciferase activity was measure and normalized to the co-transfected renilla control. Note that only the long form contains the predicted TEAD recognition motifs (upstream of −1 075). Data are means ± SD from triplicates in a representative experiment. (E) TEAD1 binds to the SLC7A5 promoter region. Chromatin immunoprecipitation (CHIP) with antibodies to endogenous TEAD1 (or control IgG) were carried out in 293A cells. The precipitated DNA was quantitated by real-time PCR analysis with primers specific for a promoter region (prom) or an intragenic control region (intra) of the indicated genes. CTGF (connective tissue growth factor) is a known direct target gene of the YAP-TEAD complex, and GAPDH (glyceraldehyde 3-phosphate dehydrogenase) serves as an additional negative control. Data are means ± SEM of triplicates from a representative experiment. (F) Ectopic expression of active YAP or TAZ increases SLC7A5 mRNA expression. qPCR from Y/T dbKO cells expressing doxycycline-inducible active YAP/TAZ as depicted. The cells were serum starved and doxycycline-induced for 16 h. Data are means ± SEM of triplicates from a representative experiment. (G) Ectopic expression of active YAP or TAZ increases SLC7A5 protein levels. Experiments are similar to F. * denotes antibody (sc-101199) that recognizes both YAP and TAZ. (H) SLC7A5 expression is dependent on YAP-TEAD interaction. Western blots of cell lysates from Y/T dbKO cell lines stably re-expressing YAP, or the TEAD binding-deficient YAP mutant (S94A). Note that the TEAD binding-deficient S94A YAP does not rescue the expression of SLC7A5. (I) Leucine uptake is dependent on YAP-mediated TEAD activation. H3-leucine uptake assay was carried out on Y/T dbKO cell lines with stable expression of vector control, YAP, and the TEAD binding-deficient YAP S94A mutant. Error bars are means ± SEM of triplicates from a representative experiment.
To assess whether the TEAD transcription factors directly bind to the SLC7A5 promoter in cells, we performed chromatin immunoprecipitation (ChIP) and confirmed that TEAD1 did bind to the SLC7A5 promoter region (Figure 4E). Next, we tested whether YAP/TAZ can induce SLC7A5 mRNA expression. We generated doxycycline-inducible cell lines expressing active YAP S127A or TAZ S89A (YAP/TAZ mutants lacking a key inhibitory phosphorylation site). Induction of active YAP or TAZ increased SLC7A5 mRNA and protein expression (Figure 4F and 4G). To assess whether this response is dependent on the TEADs, we utilized a YAP mutant (S94A) that is deficient in TEAD binding6,42,43. Compared to WT YAP, the TEAD binding-deficient YAP S94A mutant was unable to induce SLC7A5 expression in Y/T dbKO cells (Figure 4H). Furthermore, while reintroduction of WT YAP restored Leu uptake, reintroduction of the TEAD binding-deficient YAP S94A mutant failed to do so (Figure 4H and 4I), confirming that SLC7A5 is a direct YAP-TEAD target gene critical for Leu uptake.
YAP/TAZ act through SLC7A5 to promote cell growth
Our data suggest that YAP/TAZ induces SLC7A5 expression to increase Leu uptake, thereby activating mTORC1 to provide a growth advantage for cells under low nutrient conditions. This hypothesis predicts that decreased mTORC1 activation by Gln/Leu stimulation in Y/T dbKO cells is due to reduced SLC7A5 expression. To test this model, we reintroduced SLC7A5 back into Y/T dbKO cells and measured mTORC1 activity. As shown in Figure 5, re-expression of SLC7A5 restored mTORC1 activation in the Y/T dbKO cells (Figure 5A-5C and Supplementary information, Figure S4A-S4B), whereas re-expression of SLC3A2 did not (Supplementary information, Figure S4C). This is consistent with the fact that SLC3A2 re-expression alone fails to restore protein levels of SLC7A5 (Supplementary information, Figure S3A). This effect was specific to Gln/Leu stimulation, as no difference was observed in mTORC1 activation in response to insulin when cells were cultured in serum-free and AA-containing DMEM (Figure 5D). These results indicate that loss of SLC7A5 expression in Y/T dbKO cells is the primary reason for defective mTORC1 activation by Gln/Leu stimulation.
YAP/TAZ act through SLC7A5 to promote cell growth at low AA levels. (A) Ectopic SLC7A5 expression restores mTORC1 activation by AAs in Y/T dbKO KO cells. Y/T dbKO cells stably expressing vector or V5-tagged SLC7A5 were co-cultured. Cells were AA starved and then stimulated with Gln/Leu in the absence (upper panels) or presence (lower panels) of rapamycin. Note that SLC7A5 re-expression (marked by positive V5 staining, red) rescues pS6 stimulation upon Gln/Leu stimulation. See Supplementary information, Figure S4A and S4B for larger field of cells. Scale bar, 20 μm. (B) Ectopic SLC7A5 expression rescues mTORC1 activation by AAs. Experiments are similar to A. mTORC1 activation is determined by western blotting for phosphorylation of S6K and 4E-BP1. (C) Re-expression of SLC7A5 rescues S6K activation. Quantification of pS6K signals after 10 min of Leu stimulation from experiments as in B. Error bars are means ± SEM of triplicates. (D) SLC7A5 does not alter insulin-induced mTORC1 activation. Cells were serum starved and then stimulated with 100 nM insulin for the times indicated. Tor denotes 1-h pretreatment with Torin prior to insulin stimulation. (E) YAP/TAZ act through SLC7A5 to control cell size. The indicated cell lines were cultured for 36 h in normal DMEM (AA+), 10% concentration of all AAs, or normal concentration of all AAs except leucine, which was 1%. The relative cell size was determined by FACS and normalized to WT cells grown in full media. Data depicted is the average normalized cell size from four replicates. Error bars are means ± SEM from a representative experiment. (F) Y/T dbKO cells ectopically expressing SLC7A5 outgrow Y/T dbKO cells at low AA concentrations. Representative confocal images of cells labeled for V5 and stained with DAPI. Y/T dbKO cells expressing vector control or V5-tagged SLC7A5 were seeded at low density and grown for 144 h with medium change every 24 h, whereafter cells were seeded into LabTek chambers and processed for immunofluorescence on the following day. Note that SLC7A5 re-expressing cells outgrow Y/T dbKO control cells at low AA (middle row) and especially low leucine concentrations (bottom row), but not at full AA concentrations (top row). See Supplementary information, Figure S3D for pure cell cultures. Scale bar, 50 μm. (G) Expression of SLC7A5 confer cell growth advantages at low AA concentrations. Cell competition assay in co-culture of Y/T dbKO cells expressing either empty vector or V5-tagged SLC7A5 grown under different conditions for 144 h as in F. Medium was changed every 24 h to keep serum concentration within a similar range. The ratio of V5-expressing cells at the end of the experiment was quantified using FACS and data depicted are means ± SEM from a representative experiment.
A well-defined function of mTORC1 is to promote cell growth, as measured by an increase in cell size27,30,44. We next investigated whether YAP/TAZ or SLC7A5 have any effect on cell size. The Y/T dbKO cells were slightly smaller than the WT cells when cultured under nutrient sufficiency (in complete DMEM; Figure 5E). Because LAT1 is a high-affinity amino acid transporter, we examined cell size at reduced amino acid concentrations. As expected, reduction of total amino acid to 10%, or reduction of Leu to 1% of normal DMEM resulted in decreased cell size, potentially due to partial inhibition of mTORC127,30. Interestingly, when cultured under reduced amino acid concentrations, the Y/T dbKO cells showed a dramatic decrease in cell size (Figure 5E), consistent with decreased mTORC1 activity. Furthermore, re-expression of SLC7A5 significantly rescued cell size in the Y/T dbKO cells. It is worth noting that the re-expression of SLC7A5 could not completely rescue the cell size difference between Y/T dbKO and WT cells, possibly due to other YAP/TAZ target genes involved in cell growth regulation1,2.
We next examined whether the ability to take up AAs would provide a growth advantage for cells expressing SLC7A5. We co-cultured equal numbers of Y/T dbKO cells expressing either empty vector or SLC7A5 at different AA concentrations. Cells were counted six days later. The SLC7A5-expressing Y/T dbKO cells clearly outgrew the control cells at low AA concentrations, whereas the difference was much less pronounced when cultured under nutrient sufficiency (Figure 5F, 5G and Supplementary information, Figure S4B). Expression of SLC7A5 therefore confers growth advantages at low AA levels, and since expression of SLC7A5 is dependent on YAP/TAZ, these transcriptional co-activators thereby confer growth advantage under nutrient-limiting conditions.
Discussion
The Hippo pathway acts through YAP and TAZ to limit tissue growth by regulating cell proliferation and apoptosis2. mTOR promotes cell growth by stimulating synthesis of primary building blocks, such as nucleotides, lipids, and proteins, and by inhibiting autophagy45,46. Nutrient scarcity is a common phenomenon in biological processes. Cells and organisms must adjust to fluctuating nutrient levels, especially during development and regeneration2,47,48. Crosstalk between potent cellular growth pathways, such as the Hippo and mTOR pathways, on multiple levels is crucial to maintain homeostasis1,45,49,50,51,52. This report shows that cells with high YAP/TAZ activity gain an enhanced ability of acquiring nutrients to support biosynthesis and growth. This is mediated through the YAP/TAZ-induced expression of the high-affinity amino acid transporter LAT1; YAP/TAZ directly increase transcription of SLC7A5, thus indirectly leading to accumulation of SLC3A2 and, therefore, high LAT1 activity.
Hyperactive YAP/TAZ2 and high LAT153 expression have been observed in a range of cancers, suggesting that they may contribute to tumor development. Because LAT1 is a high-affinity amino acid transporter and solid tumors often experience nutrient limitation due to insufficient blood supply, elevated LAT1 in tumor cells, potentially due to mutations in the Hippo pathway, will gain the upper hand in competing with surrounding wild-type cells for nutrients. Therefore, targeting either YAP/TAZ and/or LAT1 (CD98) may be an attractive therapeutic target in treating YAP/TAZ-driven cancers2,4,50,53,54,55,56,57,58,59. The finding that YAP/TAZ regulate SLC7A5 expression and LAT1 activity to activate mTORC1 is consistent with a recent report50, and together these studies represent an important step in understanding how YAP/TAZ target genes promote cell growth and how these potent growth regulatory pathways are interconnected.
Materials and Methods
Image acquisition
A Zeiss 710 confocal microscope utilizing a Plan-Apochromat 63×/1.4 Oil DIC M27 objective was used to acquire images. Cells were grown on fibronectin-coated LabTek chambers or coverslips, and after experimental procedures cells were PFA-fixed and mounted with prolong gold anti-fade reagent with DAPI (P36931, Life Technologies). For quantification of phosphor-S6 (pS6), laser powers were adjusted so that rapamycin-treated cells appeared negative; hereafter the same acquisition settings were used for comparable experiments. Quantification was carried out by eye and scored as fraction of pS6-positive cells per image, only fields of view with more than 6 cells per cell type quantified were included in the quantification, and in general 20 cells on average were present in one image. Scatter plots represent fraction of cells that are pS6 positive. To address antibody specificity of YAP and TAZ in confocal images, samples were prepared in parallel to ensure the exact same incubation time and antibody solutions. In images acquired for the cell competition assay (see separate section), cells were grown on plastic (with no extra coating) in experimental medium (with medium change every 24 h) for six days, whereafter cells were trypsinized and some were reseeded into LabTek chambers and processed for immunofluorescence. In general, images that are directly compared were acquired with the same microscope and laser settings. Cells with heavily condensed chromosomes as identified in the DAPI channel, and therefore undergoing either mitosis or apoptosis, were excluded in all image-based quantification.
Cell culture and AA experiments
All cell lines were cultured under an atmosphere of 5% CO2 at 37 °C. Generation of RagA/B KO 293A cells were described elsewhere24. Concentration of Gln and Leu as 1×, are the concentrations present in DMEM high-glucose media #11965. 1× Gln equals 4 mM, 1× Leu equals 0.801 mM. Amino acid-free medium was made in house following the Invitrogen (#11965-092) high-glucose DMEM recipe, but omitting all AAs. All experiments with AA starvation and stimulation contained 10% dialyzed FBS (#26400-036 from Invitrogen) unless otherwise indicated. In experiments compared to full AA-containing medium, 10% dialyzed FBS was likewise used.
Generation of stable cell lines
Stable cells expressing pQCXIH-Myc-YAP, and pQCXIH-myc YAP s94A were generated by retroviral transduction and selected with hygromycin, and these plasmids have previously been described6. V5-tagged SLC7A5 and V5-tagged SLC7A7 were acquired from DNASU60. HsCD00042452 (SLC7A5), HsCD00040795 (SLC7A7) and HsCD00513893 (SLC3A2) are inserted into pDonR221 and were in-house via gateway cloning inserted into the lentiviral pLEX307 destination vector, generating C-terminally V5-tagged transporters for mammalian expression. Stable cells were obtained through puromycin selection. Empty pLEX307 was used as a control. Retrovirus expressing pLKO-based vectors with shRNA targeting SLC7A5 (TRCN0000043009, TRCN0000043008, TRCN0000043010, TRCN0000333531) were acquired from Sigma, and shRNA control described previously35 was prepared. Cells were transduced with the retrovirus and expressing cells were puromycin selected.
For generation of tetracycline-inducible expression system, cells were infected with a retrovirus encoding tetracycline repressor (TetR; pRetroX-Tet-On; Invitrogen) for 2 days. The cells were then incubated with G418 (300 μg/ml) for selection and amplification of neomycin-resistant cells. The TetR-expressing cells were then infected with retroviruses encoding YAP (s127A), TAZ (s89A) (or pRetroX-Tight-Pur empty vector for control). Forty-eight hours after infection, the cells were selected with G418 (300 μg/ml) and puromycin (4 μg/ml) in culture medium. For induction of YAP/TAZ expression, the cells were treated with 0.5 μg/ml doxycycline (dox) for the indicated times. Prior to induction, cells were grown for at least 4 days in medium containing Tet System Approved Serum (Fisher Scientific) to avoid leakage.
Transfection of plasmid DNA was performed with PolyJet DNA In Vitro Transfection Reagent (SignaGen Laboratories) according to manufacturer's instructions.
Antibodies and reagents
Antibodies (abs) from Cell signaling used in this study are against SLC3A2 (also known as 4F2hc) (D603P), against SLC7A5 (also known as LAT1) (53475), against phosphor-4E-BP1 (Thr37/46) (236B4), against total 4E-BP1 (9452), against total S6 (5G10), against phosphor S6 (D57.2.2E and 5364), against p127 YAP (D9W23), against phosphor p70 kinase (9234L), against total p70 kinase (9202), against LATS1 (C66B5), against MYC (9B11), against TAZ (V386), against RagA (4357). YAP (EP1674Y; human specific) ab was from Abcam. Antibodies against HSP90 (610418) and TEAD1 (610922) were from BD Biosciences. V5 antibody was from Life Technologies (R960-25). Vincullin (V9131) was from Sigma. Antibodies against GAPDH (sc-25778), Cyr61 (sc-13100) and YAP (sc-101199/63.7) were all from Santa Cruz Biotechnology. Santa Cruz ab (sc-101199/63.7) is raised against a YAP peptide in the region 50-165. This Ab also recognize TAZ, see description of generation of YAP/TAZ dbKO cells (Supplementary information, Figure S1B and S1C). The peptide used to generate the EP1674Y Abcam antibody is in the region aa 360-400 (exact position is not available as this information is proprietary). Fluorescent secondary abs used were mouse and rabbit Alexa Fluor 488 and 594, both raised in goat and obtained from Molecular Probes/Invitrogen. Alexa Fluor 488 Phalloidin was likewise from Molecular Probes. Fibronectin was from Sigma (F-1141).
Radioactive leucine uptake experiment
Cells were seeded onto fibronectin-coated 24-well plates, and briefly washed once in Na2+ free HBSS prior to the start of experiment, followed by 10-min incubation in Na2+ free HBSS. Na2+ free HBSS is 137 mM Choline Chloride, 5.3 mM KCl, 1.3 mM CaCl2, 0.49 mM MgCl2, 0.41 mM MgSO4, 0.35 mM K2HPO4, 0.44 mM KH2PO4, 4.2 mM KHCO3, 5.6 mM D-Glucose adjusted to pH 7.4. The cells were hereafter allowed to take up 1 mM leucine supplemented with titriated leucine for 3 min, rinsed three times with ice-cold Na2+ free HBSS and lysed with 200 μL 0.1 M NaOH. 150 μL of the mixture was taken into scintillation fluid (optiphase supermix, PerkinElmer) in LSC vials (PerkinElmer) and measured on a liquid scintillation analyzer. Each time point measurement was carried out in triplicates.
qPCR and primers used
Cells were harvested for RNA extraction using RNeasy Plus mini kit (QIAGEN). RNA samples were reverse-transcribed to complementary DNA (cDNA) using iScript reverse transcriptase (Bio-Rad). qRT-PCR was performed using KAPA SYBR FAST qPCR kit (Kapa Biosystems) and the 7300 real-time PCR system (Applied Biosystems). Expression levels were normalized to HPRT.
qPCR primers used were: 5′-AGAATGTCTTGATTGTGGAAGA-3′ and 5′-ACCTTGACCATCTTTGGATTA-3′ for HPRT; 5′-GGAAGGGTGATGTGTCCAATC-3′ and 5′-TAATGCCAGCACAATGTTCCC-3′ for SLC7A5; 5′-CCAATGACAACGCCTCCTG-3′ and 5′-TGGTGCAGCCAGAAAGCTC-3′ for CTGF; 5′-AGCCTCGCATCCTATACAACC-3′ and 5′-TTCTTTCACAAGGCGGCACTC-3′ for CYR61; 5′-GGGAGCAGAAGTATTTGGAGGAACG-3′ and 5′-GAATGTCGGATGAGAGTGGTGTCAC-3′ for AMOTL2.
ChIP assay
Chromatin DNA was digested with micrococcal nuclease (MNase). Immunoprecipitation reactions with either TEAD1 (BD, 610922) or mouse IgG control were carried out with chromatin extracts overnight at 4 °C. Precipitated DNA was quantitated by real-time PCR analysis. All ChIP signals were normalized to the input (labeled as percentage of input on the vertical axis) as described35 (see Supplementary information, Data S1 for a more detailed protocol).
Generation of knockout cells
CRISPR/Cas9 system was utilized with the following guide sequences:
slc7a5 set #1:
caccgcgtggggaccattatcggct
aaacagccgataatggtccccacgc
slc7a5 set #2:
caccgacggcgtggccatcatcgtg
aaaccacgatgatggccacgccgtc
yap:
caccgcatcagatcgtgcacgtccg
aaaccggacgtgcacgatctgatgc
wwtr1 (taz) set #1:
caccgtgtctaggtcctgcgtgacg
aaaccgtcacgcaggacctagacac
wwtr1 (taz) set #2:
caccgatcacttgctgcccaggcgg
aaacccgcctgggcagcaagtgatc
The above designed oligos were annealed and cloned into pSpCas9(BB)-2A-Puro (PX459; Addgene plasmid #48139). Following sequence verification, these oligos were transfected into 293a cells utilizing Polyjet. On the following day puromycin was added to the transfected cells for three days. Hereafter cells were allowed to recover in full medium without puromycin for two days. Cells were then trypsinized, washed with PBS containing 5% FBS, run through a cell strainer, and re-suspended in fluorescence-activated cell sorting (FACS) buffer (PBS, 5 mM EDTA, 2% FBS). Cells were single cell sorted by FACS (UCSD; Human Embryonic Stem Cell Core, BD influx) into a 96-well plate containing DMEM with 20% FBS, 1× Sodium pyruvate (Gibco) and 50 μg/ml penicillin/streptomycin. The cells were expanded and screened, and knockout clones were selected by immunoblot analysis.
Luciferase assay and promoter cloning
A 2 000 bp fragment (long) or 1 170 bp (short) of human genomic DNA containing the promoter region of SLC7A5 was cloned into the pGL3-Basic vector. HEK293A YAP/TAZ dbKO cells were cotransfected with either pGL3 empty vector, or pGL3 SLC7A5 luciferase reporter in combination with pCMV Flag-YAP, and Renilla plasmids as indicated. Luciferase assay was performed 30 h after transfection using the Dual Glo luciferase system (Promega) and carried out on a TECAN Infinite F200 plate reader according to the manufacturer's recommendations. All luciferase activities were normalized to Renilla.
Western blotting
Cells were lysed in sample buffer with β-mercaptoethanol (reducing), or without β-mercaptoethanol (non-reducing). Cell lysates were in general prepared in reducing lysis buffer if not specifically highlighted in the figure legend. Blotted PVDF was incubated in blocking solution (5% dry milk containing TBST) for 30 min, followed by 3× min of brief wash in TBST, and incubation with primary antibody in TBST supplemented with 5% BSA overnight, followed by brief wash with TBST, whereafter the membrane was incubated in 5% milk containing TBST with the appropriate HRP-conjugated antibodies for 1 h. Western blots were developed using Millipore Immobilon Western Chemiluminescent HRP mix (WBKLS0500) onto Blue Devil autoradiography films from Genesee Scientific. Quantification of western blots for pS6K was carried out in Image J/Fiji and the values were normalized to relative total S6K levels.
FACS for cell competition assay and relative cell size determination
In total 120 000 cells were seeded in a cell competition assay into a 12-well plate. A 1:1 mixture of Y/T dbKO cells expressing an empty vector, or re-expressing V5-tagged SLC7A5 were seeded. Pure lines were included to allow for correct gating on the FACS and control for cell line purity (all cells in the V5-expressing cell line were expressing V5). Cells were seeded and allowed to settle overnight. On the following day medium was changed into experimental medium, and this medium change was repeated every 24 h to allow for serum replenishment. After 144 h, cells were trypsinized and processed for FACS using antibodies recognizing V5 and by labeling with Alexa 488. In total 10 000 events were counted and cells were gated to exclude duplets. This allowed for calculating the percentage of V5-positive cells in the mixed cell cultures after growth in different media. To determine the relative cell size by FACS, cells were seeded at low density to ensure that they were subconfluent at the end of 48 h, and medium was changed after 24 h to ensure that serum was within a constant range.
Cells were prepared for FACS, and gated against debris and for doublet discrimination. The average relative cell size per sample was compared to WT with 100% AA and plotted in Prism.
References
Hansen CG, Moroishi T, Guan KL . YAP and TAZ: a nexus for Hippo signaling and beyond. Trends Cell Biol 2015; 25:499–513.
Moroishi T, Hansen CG, Guan KL . The emerging roles of YAP and TAZ in cancer. Nat Rev Cancer 2015; 15:73–79.
Varelas X . The Hippo pathway effectors TAZ and YAP in development, homeostasis and disease. Development 2014; 141:1614–1626.
Johnson R, Halder G . The two faces of Hippo: targeting the Hippo pathway for regenerative medicine and cancer treatment. Nat Rev Drug Disco 2014; 13:63–79.
Piccolo S, Dupont S, Cordenonsi M . The biology of YAP/TAZ: hippo signaling and beyond. Physiol Rev 2014; 94:1287–1312.
Zhao B, Ye X, Yu J, et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev 2008; 22:1962–1971.
Ota M, Sasaki H . Mammalian Tead proteins regulate cell proliferation and contact inhibition as transcriptional mediators of Hippo signaling. Development 2008; 135:4059–4069.
Vassilev A, Kaneko KJ, Shu H, Zhao Y, DePamphilis ML . TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/Yes-associated protein localized in the cytoplasm. Genes Dev 2001; 15:1229–1241.
Camargo FD, Gokhale S, Johnnidis JB, et al. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr Biol 2007; 17:2054–2060.
Zhao B, Wei X, Li W, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev 2007; 21:2747–2761.
Dong J, Feldmann G, Huang J, et al. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell 2007; 130:1120–1133.
Yu FX, Luo J, Mo JS, et al. Mutant Gq/11 promote uveal melanoma tumorigenesis by activating YAP. Cancer Cell 2014; 25:822–830.
Feng X, Degese MS, Iglesias-Bartolome R, et al. Hippo-independent activation of YAP by the GNAQ uveal melanoma oncogene through a trio-regulated rho GTPase signaling circuitry. Cancer Cell 2014; 25:831–845.
Liu G, Yu FX, Kim YC, et al. Kaposi sarcoma-associated herpesvirus promotes tumorigenesis by modulating the Hippo pathway. Oncogene 2015; 34:3536–3546.
Steinhardt AA, Gayyed MF, Klein AP, et al. Expression of Yes-associated protein in common solid tumors. Hum Pathol 2008; 39:1582–1589.
Lamar JM, Stern P, Liu H, Schindler JW, Jiang ZG, Hynes RO . The Hippo pathway target, YAP, promotes metastasis through its TEAD-interaction domain. Proc Natl Acad Sci USA 2012; 109:E2441–E2450.
Zhou D, Conrad C, Xia F, et al. Mst1 and Mst2 maintain hepatocyte quiescence and suppress hepatocellular carcinoma development through inactivation of the Yap1 oncogene. Cancer Cell 2009; 16:425–438.
Chan SW, Lim CJ, Guo K, et al. A role for TAZ in migration, invasion, and tumorigenesis of breast cancer cells. Cancer Res 2008; 68:2592–2598.
Bar-Peled L, Sabatini DM . Regulation of mTORC1 by amino acids. Trends Cell Biol 2014; 24:400–406.
Kim SG, Buel GR, Blenis J . Nutrient regulation of the mTOR complex 1 signaling pathway. Mol Cells 2013; 35:463–473.
Jewell JL, Russell RC, Guan KL . Amino acid signalling upstream of mTOR. Nat Rev Mol Cell Biol 2013; 14:133–139.
Sancak Y, Peterson TR, Shaul YD, et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 2008; 320:1496–1501.
Hara K, Yonezawa K, Weng QP, Kozlowski MT, Belham C, Avruch J . Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J Biol Chem 1998; 273:14484–14494.
Jewell JL, Kim YC, Russell RC, et al. Metabolism. Differential regulation of mTORC1 by leucine and glutamine. Science 2015; 347:194–198.
Li L, Kim E, Yuan H, et al. Regulation of mTORC1 by the Rab and Arf GTPases. J Biol Chem 2010; 285:19705–19709.
Nicklin P, Bergman P, Zhang B, et al. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell 2009; 136:521–534.
Kim DH, Sarbassov DD, Ali SM, et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 2002; 110:163–175.
Wang X, Campbell LE, Miller CM, Proud CG . Amino acid availability regulates p70 S6 kinase and multiple translation factors. Biochem J 1998; 334(Pt 1):261–267.
Christie GR, Hajduch E, Hundal HS, Proud CG, Taylor PM . Intracellular sensing of amino acids in Xenopus laevis oocytes stimulates p70 S6 kinase in a target of rapamycin-dependent manner. J Biol Chem 2002; 277:9952–9957.
Fingar DC, Salama S, Tsou C, Harlow E, Blenis J . Mammalian cell size is controlled by mTOR and its downstream targets S6K1 and 4EBP1/eIF4E. Genes Dev 2002; 16:1472–1487.
Poncet N, Mitchell FE, Ibrahim AF, et al. The catalytic subunit of the system L1 amino acid transporter (slc7a5) facilitates nutrient signalling in mouse skeletal muscle. PloS One 2014; 9:e89547.
Sinclair LV, Rolf J, Emslie E, Shi YB, Taylor PM, Cantrell DA . Control of amino-acid transport by antigen receptors coordinates the metabolic reprogramming essential for T cell differentiation. Nat Immunol 2013; 14:500–508.
Mastroberardino L, Spindler B, Pfeiffer R, et al. Amino-acid transport by heterodimers of 4F2hc/CD98 and members of a permease family. Nature 1998; 395:288–291.
Meier C, Ristic Z, Klauser S, Verrey F . Activation of system L heterodimeric amino acid exchangers by intracellular substrates. EMBO J 2002; 21:580–589.
Moroishi T, Park HW, Qin B, et al. A YAP/TAZ-induced feedback mechanism regulates Hippo pathway homeostasis. Genes Dev 2015; 29:1271–1284.
Meng Z, Moroishi T, Mottier-Pavie V, et al. MAP4K family kinases act in parallel to MST1/2 to activate LATS1/2 in the Hippo pathway. Nat Commun 2015; 6:8357.
Milkereit R, Persaud A, Vanoaica L, Guetg A, Verrey F, Rotin D . LAPTM4b recruits the LAT1-4F2hc Leu transporter to lysosomes and promotes mTORC1 activation. Nat Commun 2015; 6:7250.
Broer S, Palacin M . The role of amino acid transporters in inherited and acquired diseases. Biochem J 2011; 436:193–211.
Taylor PM . Role of amino acid transporters in amino acid sensing. Am J Clin Nutr 2014; 99:223S–230S.
Palm W, Park Y, Wright K, Pavlova NN, Tuveson DA, Thompson CB . The utilization of extracellular proteins as nutrients is suppressed by mTORC1. Cell 2015; 162:259–270.
Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL . Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol 2008; 10:935–945.
Mo JS, Meng Z, Kim YC, et al. Cellular energy stress induces AMPK-mediated regulation of YAP and the Hippo pathway. Nat Cell Biol 2015; 17:500–510.
Li Z, Zhao B, Wang P, et al. Structural insights into the YAP and TEAD complex. Genes Dev 2010; 24:235–240.
Edinger AL, Thompson CB . Akt maintains cell size and survival by increasing mTOR-dependent nutrient uptake. Mol Biol Cell 2002; 13:2276–2288.
Shimobayashi M, Hall MN . Making new contacts: the mTOR network in metabolism and signalling crosstalk. Nat Rev Mol Cell Biol 2014; 15:155–162.
Russell RC, Yuan HX, Guan KL . Autophagy regulation by nutrient signaling. Cell Res 2014; 24:42–57.
Efeyan A, Comb WC, Sabatini DM . Nutrient-sensing mechanisms and pathways. Nature 2015; 517:302–310.
Wackerhage H, Del Re DP, Judson RN, Sudol M, Sadoshima J . The Hippo signal transduction network in skeletal and cardiac muscle. Sci Signal 2014; 7:re4.
Tumaneng K, Schlegelmilch K, Russell RC, et al. YAP mediates crosstalk between the Hippo and PI(3)K-TOR pathways by suppressing PTEN via miR-29. Nat Cell Biol 2012; 14:1322–1329.
Park YY, Sohn BH, Johnson RL, et al. YAP1 and TAZ Activates mTORC1 pathway by regulating amino acid transporters in hepatocellular carcinoma. Hepatology 2015 Sep 21. doi:10.1002/hep.28223
Liang N, Zhang C, Dill P, et al. Regulation of YAP by mTOR and autophagy reveals a therapeutic target of tuberous sclerosis complex. J Exp Med 2014; 211:2249–2263.
Artinian N, Cloninger C, Holmes B, Benavides-Serrato A, Bashir T, Gera J . Phosphorylation of the Hippo pathway component AMOTL2 by the mTORC2 kinase promotes YAP signaling, resulting in enhanced glioblastoma growth and invasiveness. J Biol Chem 2015; 290:19387–19401.
Bhutia YD, Babu E, Ramachandran S, Ganapathy V . Amino Acid transporters in cancer and their relevance to “glutamine addiction”: novel targets for the design of a new class of anticancer drugs. Cancer Res 2015; 75:1782–1788.
Park HW, Guan KL . Regulation of the Hippo pathway and implications for anticancer drug development. Trends Pharmacol Sci 2013; 34:581–589.
Estrach S, Lee SA, Boulter E, et al. CD98hc (SLC3A2) loss protects against ras-driven tumorigenesis by modulating integrin-mediated mechanotransduction. Cancer Res 2014; 74:6878–6889.
Ablack JN, Cantor JM, Metz PJ, Chang JT, Ginsberg MH . Ubiquitination of CD98 limits cell proliferation and clonal expansion. J Cell Sci 2015 Oct 22. pii: jcs.178129
Feral CC, Nishiya N, Fenczik CA, Stuhlmann H, Slepak M, Ginsberg MH . CD98hc (SLC3A2) mediates integrin signaling. Proc Natl Acad Sci USA 2005; 102:355–360.
Henderson NC, Collis EA, Mackinnon AC, et al. CD98hc (SLC3A2) interaction with beta 1 integrins is required for transformation. J Biol Chem 2004; 279:54731–54741.
Mohamed AD, Tremblay AM, Murray GI, Wackerhage H . The Hippo signal transduction pathway in soft tissue sarcomas. Biochim Biophys Acta 2015; 1856:121–129.
Seiler CY, Park JG, Sharma A, et al. DNASU plasmid and PSI:Biology-Materials repositories: resources to accelerate biological research. Nucleic Acids Rese 2014; 42:D1253–D1260.
Acknowledgements
We would like to thank members of the Guan lab, especially Drs Toshiro Moroishi and Ryan Russell, for insightful discussions during the course of this study, as well as Dr Jung-Soon Mo for lending a helping hand with cell culture and the always helpful Fabian Flores for proofreading. Jesus Olvera and Cody Fine from the UC San Diego Human Embryonic Stem Cell Core Facility are acknowledged for their help and technical advice during cell sorting and FACS. The work in the Guan laboratory was supported by a National Institutes of Health (NIH) grant (CA132809 and EYO226116) to KLG. CGH was initially supported by a Postdoctoral Fellowship from the Danish Council for Independent Research (Natural Sciences). SWP is supported by a University of California at San Diego Cellular and Molecular Pharmacology training grant (T32 GM007752).
Author information
Authors and Affiliations
Corresponding author
Additional information
( Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary information
Supplementary information, Figure S1
(A) Glutamine is required for leucine to activate mTORC1. (PDF 2525 kb)
Supplementary information, Figure S2
SLC7A5 is predominantly localized to the plasma membrane. (PDF 2278 kb)
Supplementary information, Figure S3
(A) Re-expression of ectopically V5 tagged SLC3A2 in Y/T dbKO cells do not rescue SLC7A5 expression. (PDF 454 kb)
Supplementary information, Figure S4
(A) SLC7A5 rescues activation of mTORC1 by Gln/Leu in YAP/TAZ dbKO cells. (PDF 1056 kb)
Supplementary information, Data S1
Materials and Methods (PDF 239 kb)
Rights and permissions
About this article
Cite this article
Hansen, C., Ng, Y., Lam, WL. et al. The Hippo pathway effectors YAP and TAZ promote cell growth by modulating amino acid signaling to mTORC1. Cell Res 25, 1299–1313 (2015). https://doi.org/10.1038/cr.2015.140
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cr.2015.140
Keywords
This article is cited by
-
The LKB1–TSSK1B axis controls YAP phosphorylation to regulate the Hippo–YAP pathway
Cell Death & Disease (2024)
-
Amino acid metabolism regulated by lncRNAs: the propellant behind cancer metabolic reprogramming
Cell Communication and Signaling (2023)
-
Ferroptosis in lung cancer: a novel pathway regulating cell death and a promising target for drug therapy
Cell Death Discovery (2023)
-
Extracellular-matrix mechanics regulate cellular metabolism: A ninja warrior behind mechano-chemo signaling crosstalk
Reviews in Endocrine and Metabolic Disorders (2023)
-
A YAP/TAZ-TEAD signalling module links endothelial nutrient acquisition to angiogenic growth
Nature Metabolism (2022)