Abstract
Tumor suppressor PTEN regulates cellular activities and controls genome stability through multiple mechanisms. In this study, we report that PTEN is necessary for the protection of DNA replication forks against replication stress. We show that deletion of PTEN leads to replication fork collapse and chromosomal instability upon fork stalling following nucleotide depletion induced by hydroxyurea. PTEN is physically associated with replication protein A 1 (RPA1) via the RPA1 C-terminal domain. STORM and iPOND reveal that PTEN is localized at replication sites and promotes RPA1 accumulation on replication forks. PTEN recruits the deubiquitinase OTUB1 to mediate RPA1 deubiquitination. RPA1 deletion confers a phenotype like that observed in PTEN knockout cells with stalling of replication forks. Expression of PTEN and RPA1 shows strong correlation in colorectal cancer. Heterozygous disruption of RPA1 promotes tumorigenesis in mice. These results demonstrate that PTEN is essential for DNA replication fork protection. We propose that RPA1 is a target of PTEN function in fork protection and that PTEN maintains genome stability through regulation of DNA replication.
Similar content being viewed by others
Introduction
Cell proliferation requires precise ordering of numerous processes that ensure accurate propagation of genomic information and preservation of genomic integrity1,2, and accuracy of DNA replication is essential to genomic stability2. DNA replication machinery must accomplish accurate duplication of genomic information despite numerous obstacles of both intracellular and extracellular origins that can cause replication stress-a complex phenomenon manifested as a slowing or stalling of replication fork progression and/or DNA synthesis3. Replication stress may result from oncogene activation and significantly affects genome stability, cell survival and pathogenesis of human disease, including carcinogenesis4. Following replication stress, replication protein A5 binds to the single-strand DNA lying adjacent to newly replicated double-stranded DNA that has stalled for the purpose of protecting this single naked strand2, preparing cells to overcome stress. Deregulation of DNA replication occurs when cells are unable to successfully overcome stress, and may give rise to uncontrolled initiation and fork collapse thus promoting DNA damage and genomic instability3. Hence, protection of replication from stress is critical for the maintenance of genomic integrity and prevention of tumorigenesis.
PTEN is one of the most frequently mutated genes in human cancer6. Its canonical tumor suppressor function is dephosphorylation of phosphoinositide-3,4,5-triphosphate, thus suppressing the PI3K/AKT pathway that stimulates cell growth and survival7. PTEN also exerts phosphatase activity independent-tumor suppressor activities in the nucleus, where it plays a fundamental role in the maintenance of chromosomal stability through its physical interaction with centromeres and the control of DNA repair8,9,10.
RPA is a major eukaryotic single-strand DNA-binding protein complex. It consists of three subunits, RPA1 (RPA70), RPA2 (RPA32), and RPA3 (RPA14). RPA is essential for DNA replication, telomere maintenance, DNA recombination, DNA repair, and activation of DNA damage checkpoints11,12, and has recently been shown to be a part of the replication fork protection complex13. Sufficient level of free RPA is essential for the maintenance of genomic integrity14, suggesting that RPA plays a significant role in cancer suppression.
Ubiquitination is a reversible posttranslational modification. Ubiquitin (Ub)-substrate isopeptide bonds can be cleaved by deubiquitinases (DUBs). Five DUB families have been identified including ovarian tumor proteases (OTUs)15. OTUB1 is an OTU family DUB cysteine protease highly specific for cleaving Lys48-linked polyubiquitin chains, which targets proteins for proteasomal degradation16,17,18,19.
In this study, we demonstrate that PTEN plays an essential role in DNA replication fork protection and thus maintains genome stability under replication stress. PTEN is located at replication sites, and physically interacts with RPA1 as well as OTUB1, which regulates RPA1 stability. In addition, expression of PTEN and RPA1 shows strong correlation in colorectal cancer. Consistent with these findings, heterozygous disruption of RPA1 by homologous recombination promotes tumorigenesis in mice. Based on these data we propose a novel mechanism of tumor suppression whereby PTEN regulates RPA1 protein stability and protects the DNA replication fork to maintain genomic stability against replication stress.
Results
PTEN protects stalled replication forks in a phosphatase-independent manner
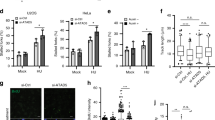
To determine whether PTEN is involved in DNA replication, DNA fiber analysis was carried out to evaluate replication perturbation genome-wide at single-molecule resolution20. Stability of nascent replication tracts was monitored in wild-type and Pten null mouse embryonic fibroblasts (MEFs; Pten+/+ and Pten−/− MEFs; Figure 1A). The median length of 5-iodo-2′-deoxyuridine (IdU) tracts was slightly shortened with hydroxyurea (HU) treatment in Pten+/+ MEFs, indicating that the integrity of stalled forks is not significantly compromised during prolonged replication stress. In contrast, nascent IdU tracts showed substantial shortening in association with replication fork stalling in Pten−/− MEFs as compared with unperturbed replication or with perturbed Pten+/+ MEFs tracts (Figure 1A). This raised the possibility that PTEN functions to protect nascent strands on stalled replication forks.
PTEN protects stalled replication forks and suppresses genomic instability. (A) Experimental design of the DNA fiber assay. Lengths of nascent replication tracts labeled with IdU were measured by DNA spreading after 5 h of replication stalling with or without HU treatment. IdU tracts in Pten−/− MEFs show significant strand shortening with HU treatment as compared with Pten+/+ MEFs. IdU tract lengths reflect replication fork stability. (B) IdU tract lengths in PTEN+/+ and PTEN−/− HCT116 cells with or without HU treatment. (C, D) IdU tract lengths in PTEN+/+ and PTEN−/− HCT116 cells, as well as in Pten+/+ and Pten−/− MEFs with different exposure times to HU treatment. (E) Measurement of IdU tracts in PTEN−/− HCT116 cells untransfected or transfected with Flag-HA-PTENWT, PTEN mutants with or without HU treatment. PTENWT and phosphatase activity dead mutants of PTENC124S and PTENG129E were tested for the ability to rescue stalled DNA replication forks caused by PTEN deletion. (F) CldU tract lengths in PTEN+/+ and PTEN−/− HCT116 cells upon HU treatment. CldU tracts represent continuous tracts formed during HU treatment. (G) Gap lengths between IdU tracts prior to HU treatment and CldU tracts after HU treatment in PTEN+/+ and PTEN−/− HCT116 cells. Gap lengths indicate the efficiency of blocking the restart. (H) Diagram of gap lengths between IdU tracts before HU treatment and CldU tracts after HU treatment (leading strand) and CldU tract lengths with HU treatment (lagging strand) showing there is asymmetric replication fork movement in PTEN−/− HCT116 cells. (I) Chromosomal aberrations measured by metaphase chromosome spreads in PTEN+/+ HCT116 and PTEN−/− HCT116 cells with or without HU treatment. Representative images of chromosomal aberrations in metaphase chromosomes are shown. Cells with PTEN deletion show genomic instability under replication stress. All data are presented as means ± SEM and analyzed by unpaired t-test. **P< 0.01**P< 0.001.
The requirement of PTEN for protection of stalled replication forks was further evaluated in human cells with a DNA fiber assay. Normal human fibroblasts (NHF3) were infected with PTEN shRNA, which significantly reduced the PTEN level (Supplementary information, Figure S1B), or scrambled shRNA lentivirus. PTEN+/+ and PTEN−/− HCT116 colon cancer cell lines were also examined (Figure 1B), in which we used BRCA2+/+ and BRCA2+/− HCT116 cells generated by CRISPR (Supplementary information, Figure S1C) as controls as BRCA2 is involved in replication fork protection20. As shown in Supplementary information, Figure S1A, NHF3 cells infected with PTEN shRNA lentivirus (Supplementary information, Figure S1B) resulted in a decrease in IdU tract lengths under HU-induced replication stress. IdU tracts were also shorter in HU-treated PTEN−/− HCT116 cells (Figure 1B), which is similar to the phenotype observed in BRCA2+/− HCT116 cells (Supplementary information, Figure S1C). To further analyze the features of replication tract shortening during stalling, we monitored the nascent strands during the course of HU treatment. As expected, in both MEFs and HCT116 cells, IdU tracts in PTEN null cells became progressively shortened during HU treatment compared with those in PTEN normal cells (Figure 1C and 1D). We also noticed that DNA tracts from cells lacking PTEN were slightly shortened under normal condition as compared with PTEN+/+ cells (Figure 1A and 1B). These observations argue that there is a critical requirement for PTEN in the protection of stalled replication forks. Since PTEN phosphatase activities (lipid and/or protein phosphatase) are important in tumor suppression, we introduced plasmids expressing PTENWT, PTENC124S (both lipid and protein phosphatase-dead mutant) and PTENG129E (lipid phosphatase-dead mutant) into PTEN−/− HCT116 cells and monitored the nascent tracts. Surprisingly, no significant difference was found between wild type PTEN and the mutants: tract shortening was rescued by PTEN and all mutants tested (Figure 1E). We then tested the efficiency of a classic PI3K inhibitor, LY29400221, in rescue of the shortened tracts in Pten−/− MEFs. LY294002 treatment showed no effect on defective replication protection in Pten−/− MEFs (Supplementary information, Figure S1D). This indicates that PTEN protects replication forks in a phosphatase-independent manner.
PTEN suppresses strand uncoupling and maintains genome stability under replication stress
We further evaluated the involvement of PTEN in replication recovery. We analyzed PTEN−/− HCT116 cells recovering from HU-induced replication stress using flow cytometry and the γ-H2AX foci formation assay, and found they exhibited a slower S phase progression after release from HU treatment, as compared with control cells (Supplementary information, Figure S1E and S1F). We also measured CldU tract lengths after HU treatment and found that PTEN+/+ and PTEN−/− HCT116 cells showed no significant difference in CldU tract lengths (Supplementary information, Figure S1G and S1H). In addition, IdU and CldU tracts in PTEN−/− cells were partially continuous after an intervening HU treatment, similar to the PTEN+/+ cells (Supplementary information, Figure S1I-S1L), suggesting that PTEN may not influence the replication restart process. In contrast, the continuous CldU tracts formed during HU treatment in PTEN−/− cells were longer than those in PTEN+/+ cells (Figure 1F), and at the same time, gap lengths observed between two types of labeling in PTEN null cells were shorter as compared to PTEN normal cells (Figure 1G), leading to a significant difference between the continuous tracts and gaps, and indicating that there is asymmetric replication fork movement (Figure 1H). Taken together, these data suggest that PTEN is mainly involved in fork protection and is partially involved in suppressing the uncoupling of leading and lagging strands.
We next evaluated the role of PTEN in the maintenance of genomic stability in HCT116 cells under DNA replication stress induced by HU treatment. Metaphase spreads made from cells immediately after HU exposure showed elevated levels of chromosomal aberrations in PTEN−/− HCT116 cells as compared with PTEN+/+ HCT116 cells (Figure 1I), consistent with the findings in our previous work8. Chromosome breaks in PTEN−/− HCT116 cells increased markedly from an average of 1.45 to 2 aberrations per cell upon stress (Figure 1I, lane 2 vs lane 4), whereas only a moderate elevation from 1.067 to 1.225 was observed in PTEN+/+ HCT116 cells (Figure 1I, lane 1 vs lane 3). We also used fluorescence in situ hybridization (FISH) to confirm the association of chromosomal structure changes with HU treatment in PTEN+/+ and PTEN−/− HCT116 cells. Centromeres were specifically labeled with fluorescent Cy3-labeled peptide nucleic acid (PNA) probes against centromeric sequences, and chromosomes were stained with DAPI (Supplementary information, Figure S1M). Magnifications of the selected areas in the metaphase spreads (Supplementary information, Figure S1Mb1 and S1Md1-d3; DAPI and Cy3 costained with corresponding inverted grayscale) are provided for enhanced visualization of abnormal chromosomal structures. As expected, chromosome breakage was increased in PTEN−/− HCT116 cells but not in PTEN+/+ HCT116 cells after HU treatment. Thus, replication stress induced by HU selectively causes genomic instability in PTEN defective cells.
PTEN physically interacts with RPA1 and is located at DNA replication sites
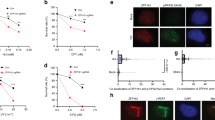
To further explore the mechanism by which PTEN protects stalled DNA replication forks, exogenous S-tagged PTEN was pulled down and PTEN-associated proteins were identified by mass spectrometry (MS) analysis (Figure 2A, lane 1). Among hits was the single-strand DNA-binding protein RPA1. RPA1 is known to function in many processes involving DNA metabolism11, thus raising the possibility that it is functionally associated with the DNA replication protection activity of PTEN. PTEN peptides were also identified in a reciprocal pull-down of exogenous S-tagged RPA1 by MS (Figure 2A, lane 3), indicating that PTEN may physically interact with RPA1. Confirming these MS results, RPA1 was detected in an anti-PTEN immunoprecipitated HCT116 cell lysate (Figure 2B) and, conversely, PTEN was detected in an anti-RPA immunoprecipitated cell lysate (Figure 2C). Immunofluorescence also showed colocalization of PTEN and RPA1 in cells (Supplementary information, Figure S2A).
PTEN binds to RPA1 C-terminus and is located at DNA replication sites. (A) In vivo S-tag pull-down analysis. Whole cell extracts from 293T cells transfected with S-tagged-PTEN, S-tagged-RPA1, or S-tagged-mock were immunoprecipitated with s-protein beads followed by mass spectrometric peptide sequencing. Both RPA1 and PTEN were found in reciprocal pull-down assays. OTUB1 was found in the PTEN and RPA1 pull-down lists. (B) In vivo binding of PTEN and RPA1. PTEN physically associates with RPA1. Lysates of HCT116 cells were immunoprecipitated with an anti-PTEN monoclonal antibody and subjected to western blotting with an anti-RPA1 antibody. (C) Reciprocal examination of the physical interaction between PTEN and RPA1. RPA1 immunoprecipitates were subjected to western blotting using anti-PTEN antibody. (D) Comparison of conventional (EPI) and STORM images of co-localization of DNA replication sites and PTEN with or without HU treatment (d3 and d4). More specific views are shown in Supplementary information, Figure S2B. STORM images of replication sites with PCNA (without HU treatment; d1) and RPA1 (with HU treatment; d2) are shown as positive control. (E, F) In vivo S-tag pull-down analysis for mapping the PTEN binding domain of RPA1. Different domains of RPA1 with S-tag were used for in vivo binding assays with Flag-tagged full-length PTEN. Various RPA1 domains are shown in the diagram (E). PTEN binds to the RPA1 C-terminal region.
Based on the interaction of PTEN and RPA1, we hypothesized PTEN may localize at replication sites. To accurately define the location of PTEN and replication sites, we used stochastic optical reconstruction microscopy or STORM22. We labeled ongoing DNA replication sites with EdU and treated Hela cells with or without HU. Strikingly, at 100 nm super resolution, PTEN was found in both conditions at the DNA replication sites delineated by EdU labeling (Figure 2D d3-d4 and Supplementary information, Figure S2B), where positive controls of PCNA (without HU treatment) and RPA1 (after HU treatment) also localized (Figure 2D d1-d2). These data suggest that PTEN is located at DNA replication sites.
A series of truncated or internally deleted RPA1 mutants were generated to further characterize PTEN-RPA1 interaction (Figure 2E). Flag-PTEN was brought down through immunoprecipitation by wild-type RPA1, and by all RPA1 mutants except the D5 mutant lacking the C-terminal domain (C domain, Figure 2F). These results suggest that RPA1 binds to PTEN via its C domain. This came as a surprise, as the C domain is not a canonical RPA1 protein–protein interaction domain11. To provide a structural basis for the physical association of PTEN and the C-terminus of RPA1, in silico docking analysis was used to model the PTEN/RPA complex. Since RPA functions as a trimer of RPA1/RPA2/RPA3, a molecular structure consisting of RPARPA1(439-616)/RPA2(43-171)/RPA323 was docked with the PTEN crystal structure24, yielding a single high probability25 PTEN/RPA complex model within a distance of 3.0 Å (Supplementary information, Figure S2C; PTEN is represented by cyan and RPA1 by salmon).
The binding of RPA1 with PTEN was next evaluated, and PTEN was found to bind RPA1 mainly through its phosphatase domain both in vitro and in vivo (Supplementary information, Figure S2D-S2F). Moreover, as revealed by in silico docking analysis, the PTEN phosphatase domain possesses a strong binding capacity for the C-terminus of RPA1 (Supplementary information, Figure S2C).
RPA1 deletion confers a phenotype with stalling of replication forks similar to PTEN deletion
RPA1 is an important single-strand DNA-binding protein, and DNA fiber assay was therefore used to evaluate RPA1 function in replication fork protection. NHF3 cells were infected with RPA1 shRNA and scrambled shRNA lentivirus as detailed in Supplementary information, Figure S3A prior to the assay. RPA1 deficient cells contained shortened IdU tracts representing unprotected stalled replication forks similar to those found in cells lacking PTEN (Supplementary information, Figure S3B, lane 4 vs lane 3 and Supplementary information, Figure S1A). To further evaluate the importance of RPA1 in the protection of stalled DNA replication forks, transcription activator-like effector nucleases (TALENs) system was used to generate RPA1 somatic knockout in HCT116 cells (Supplementary information, Figure S3C). Knockout efficiency was confirmed by DNA sequencing and western blot comparing protein expression between heterozygous knockout clones and wild-type sibling clones, which showed a significantly reduced intracellular RPA1 protein level as the result of TALEN-introduced mutation (Supplementary information, Figure S3D). The results from DNA fiber analysis in HCT116 cells (Figure 3A) corroborate those in NHF3 cells (Supplementary information, Figure S3B). Nascent IdU tracts in RPA1+/− HCT116 cells underwent shortening during replication fork stalling as compared with RPA1+/+ HCT116 cells, similar to that found in PTEN−/− HCT116 cells (Figure 3A, lanes 5 and 6 vs lanes 2 and 3 and Figure 3B). To verify the function of RPA1 in the maintenance of genomic stability under DNA replication stress, metaphase spreads of RPA1+/− and RPA1+/+ HCT116 cells were analyzed. As expected, there were significantly elevated levels of chromosomal aberrations in RPA1+/− cells upon HU treatment as compared with RPA1+/+ cells, similar to what was observed in PTEN−/− cells (Figure 3C and 1I). These findings raised the possibility that PTEN and RPA1 collaboratively protect stalled DNA forks and maintain genomic stability under replication stress.
RPA1 is a target of PTEN function in fork protection. (A) IdU tract lengths in RPA1+/+ and RPA1+/− HCT116 cells with or without HU treatment. IdU tracts in RPA1+/− HCT116 cells show significant strand shortening with HU treatment as compared with RPA1+/+ HCT116 cells. (B) IdU tracts in RPA1+/+ and RPA1+/− HCT116 cells with differing exposure times to HU. (C) Chromosomal aberrations measured by metaphase chromosome spreads from RPA1+/+ HCT116 and RPA1+/− HCT116 cells with HU treatment (± SD, n = 40). Representative images of chromosomal aberrations of metaphase chromosomes are shown. RPA1 TALEN heterozygous knockout cells show genomic instability under replication stress. (D) Measurement of IdU tract lengths in PTEN−/− HCT116 cells transfected with S-tagged-RPA1 or S-tagged-mock with or without HU. RPA1 overexpression partially rescues insufficiently protected DNA replication forks caused by PTEN deletion. (E) Chromosomal aberrations measured by metaphase chromosome spreads from PTEN−/− HCT116 cells transfected with S-tagged-RPA1 or S-tagged-mock with HU treatment. RPA1 overexpression rescues genomic instability caused by PTEN deletion under replication stress. All data are presented as means ± SEM and analyzed by unpaired t-test. **P< 0.01**P< 0.001.
RPA1 is a target of PTEN function in fork protection in response to replication stress
The similarity in the DNA fiber phenotype and karyotype between RPA1+/− HCT116 cells and PTEN−/− HCT116 cells led us to hypothesize that RPA1 functions as a target of PTEN in stalled replication fork protection. To test this hypothesis, S-tagged wild-type RPA1 was overexpressed in PTEN−/− HCT116 cells and stability of replication forks was analyzed. As shown in Figure 3D, RPA1 overexpression largely compensated for fork instability resulting from PTEN deficiency, suggesting that RPA1 functions down stream of PTEN. To determine whether RPA1 overexpression can also reverse genomic instability in PTEN null cells, metaphase spreads were evaluated in PTEN−/− HCT116 cells with induced RPA1 overexpression. As shown in Figure 3E, the average number of chromosomal aberrations per metaphase cell increased significantly following HU treatment in comparison with the control, but it was significantly reduced in PTEN−/− HCT116 cells overexpressing RPA1 (Figure 3E). Thus, genomic instability in PTEN null cells reflected by degradation of stalled replication forks can be partially reversed by RPA1 overexpression. To further confirm the relationship between PTEN and RPA1, we overexpressed PTEN in RPA1+/− HCT116 cells and performed DNA fiber assay. As expected, PTEN overexpression failed to efficiently rescue the fork instability caused by RPA1 deletion (Supplementary information, Figure S4L). These results indicate that RPA1 acts downstream of PTEN.
PTEN promotes RPA1 protein stability through regulation of RPA1 ubiquitination
To identify the mechanism by which PTEN regulates RPA1, we first evaluated RPA1 expression in PTEN+/+ and PTEN−/− HCT116 cells. As shown in Supplementary information, Figure S4A, expression of RPA1 decreased in PTEN null cells (lane 2 vs lane 1). We then treated PTEN+/+ and PTEN−/− HCT116 cells with MG132, a proteasome inhibitor that inhibits ubiquitin-mediated proteolysis, and observed that RPA1 expression was largely rescued in PTEN−/− cells (Supplementary information, Figure S4A). Post-translational modification is recognized as a regulatory mechanism of RPA1 function, and ubiquitination is related to protein stability26,27. We therefore tested if RPA1 ubiquitination is affected in cells lacking PTEN. Indeed, there was an increase of RPA1 ubiquitination in PTEN−/− cells as compared with PTEN+/+ cells (Figure 4A, lane 8 vs lane 4). This raised the possibility that PTEN is involved in regulating RPA1 ubiquitination which modulates RPA1 protein stability. Levels of endogenous RPA1 protein were thus examined in PTEN+/+ and PTEN−/− HCT116 cells in the presence of cycloheximide (CHX), an inhibitor of protein biosynthesis in eukaryotic organisms. As seen in Figure 4B, the half-life of RPA1 protein in PTEN−/− cells was significantly shortened compared to that in PTEN+/+ cells (lanes 5-8 vs lanes 1-4). These data suggest that PTEN regulates RPA1 through regulation of RPA1 ubiquitination. To determine whether PTEN lipid and protein phosphatase activities influence RPA1 stability, RPA1 protein half-life was examined in PTEN−/− HCT116 cells overexpressing PTENWT, PTENC124S or PTENG129E. There was no difference among these mutants in the ability to rescue the half-life of RPA1 (Supplementary information, Figure S4B). In addition, LY294002 failed to rescue RPA1 protein half-life in Pten−/− MEFs (Supplementary information, Figure S4C). These data indicate that PTEN affects RPA1 protein stability independent of its phosphatase activity.
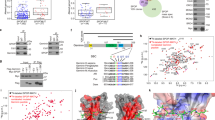
PTEN promotes RPA1 protein stability by binding and recruiting OTUB1. (A) In vivo ubiquitination assay of RPA1. PTEN+/+ and PTEN−/− HCT116 cells were co-transfected with Flag-RPA1, His-HA-ubiquitin and GFP, pulled down with Ni-beads and immunoblotted with an antibody against Flag and other antibodies as indicated. Cells were treated with MG132 (10 μM) for 12 h before collection. (B) Half-life analysis of RPA1. PTEN+/+ and PTEN−/− HCT116 cells were treated with 100 μg/ml CHX, collected at different time points and immunoblotted with antibodies against RPA1, PTEN or β-actin. Graph shows quantification of RPA1 protein levels. RPA1 protein half-life was shortened in PTEN deficient cells. (C) Chromatin fraction analysis in PTEN+/+ and PTEN−/− HCT116 cells with or without HU treatment. Asynchronized (Asyn) or HU-treated PTEN+/+ and PTEN−/− HCT116 cells were subjected to fractionation. Soluble (sol) and chromatin (chr) fractions were separated and immunoblotted with indicated antibodies. (D) Exogenous binding of PTEN, RPA1 and OTUB1. S-tagged-OUTB1 co-transfected with Flag-RPA1, Flag-PTEN, or both, or Flag-mock was pulled-down with s-protein beads. A Flag specific antibody was used to detect exogenous RPA1 and PTEN. (E) Examination of physical interaction of PTEN, RPA1, and OTUB1. OTUB1 immunoprecipitates were subjected to western blotting using anti-PTEN and anti-RPA1 antibodies. (F) In vitro binding assay examining PTEN's interaction with His-tagged-RPA1 and His-tagged-OTUB1 and both. (G) In vitro binding assay with GST-tagged full-length PTEN, various GST-tagged PTEN domains and His-tagged-OTUB1. PTEN binds to OTUB1 with its C2 domain. (H) Illustration of in silico docking analysis of the PTEN/OTUB1/RPA complex to within a distance of 3.0 Å. PTEN is represented by cyan, RPA by salmon and OTUB1 by yellow. (I) In vivo binding of OTUB1 and RPA1 in PTEN+/+ and PTEN−/− HCT116 cells. OTUB1 immunoprecipitates in PTEN+/+ and PTEN−/− HCT116 cells were subjected to western blotting using an anti-RPA1 antibody. (J) Half-life analysis of RPA1 protein. OTUB1+/+ and OTUB1−/− HCT116 cells were treated with 100 μg/ml CHX, collected at different time points and immunoblotted with antibodies against RPA1, OTUB1, and β-actin. Graph shows quantification of RPA1 protein levels. RPA1 protein expression is decreased and its half-life shortened in OTUB1 null cells. (K) In vivo ubiquitination assay of RPA1. OTUB1+/+ and OTUB1−/− HCT116 cells were co-transfected with Flag-RPA1, His-HA-ubiquitin and GFP, pulled-down with Ni-beads and immunoblotted with antibody against Flag and other antibodies as indicated. Cells were treated with MG132 for 12 h before collection. (L) IdU tract lengths in OTUB1+/+ and OTUB1−/− HCT116 cells with or without HU treatment. IdU tracts measured in OTUB1−/− HCT116 cells show significant strand shortening with HU treatment as compared with those in OTUB1+/+ HCT116 cells. Data are presented as means ± SEM and analyzed by unpaired t-test. ***P< 0.001.
The subcellular distribution of RPA1 in cells with or without replication stress was evaluated in PTEN+/+ and PTEN−/− HCT116 cells. As shown in Figure 4C, RPA1 accumulated in the chromatin fraction, and the level of accumulation was higher in PTEN+/+ cells than in PTEN−/− cells, with or without HU treatment (lanes 3, 7 vs lanes 4, 8). Furthermore, isolation of protein on nascent DNA (iPOND) analysis revealed that there was more nascent DNA colocalized with RPA1 in PTEN+/+ HCT116 cells than in PTEN−/− cells (Supplementary information, Figure S4D), suggesting that PTEN may facilitate RPA1 to accumulate on chromatin as well as at replication forks.
PTEN regulates RPA1 deubiquitination by recruiting OTUB1
Our results uncover a hitherto unknown function of PTEN in promoting the stability of its target protein. Hence, a search for DUBs that regulate ubiquitin-dependent pathways was carried out, and OTUB1 was found among the molecules pulled down by exogenous S-tagged PTEN and RPA1 (Figure 2A, lanes 1 and 3). OTUB1 is a canonical OTU family DUB19, and was therefore considered to be a potential RPA1 DUB. We sought to determine whether OTUB1 interacts with PTEN and RPA1 and functions as an RPA1 DUB. Consistent with the results of pull-down followed by MS, ectopically expressed Flag-tagged PTEN and RPA1 were present in S-tagged OTUB1 immunoprecipitate (Figure 4D), and both endogenous PTEN and RPA1 were present in endogenous OTUB1 immunoprecipitate (Figure 4E). Immunofluorescence also showed that ectopically expressed PTEN, RPA1, and OTUB1 were colocalized in PTEN−/− HCT116 cells (Supplementary information, Figure S4E). Purified PTEN bound to His-tagged OTUB1 by itself or in combination with His-tagged RPA1 under cell-free conditions (Figure 4F). PTEN truncations were next used to map the binding region with OTUB1 in vitro, and only the C2 domain of PTEN (Supplementary information, Figure S2D) bound to OTUB1 (Figure 4G, lane 4 vs lane 3). These findings argue that PTEN, RPA1, and OTUB1 may act as a complex.
To confirm the structural linkage of these molecules, in silico analysis was used to evaluate docking of elements of the OTUB1 structure28 with the PTEN crystal structure24. In the PTEN/OTUB1 complex, OTUB1 binds to the PTEN C2 domain through complimentary electrostatic charge distributions to within a distance of 3.0 Å (Supplementary information, Figure S4F; PTEN is in green and OTUB1 in yellow). We then merged PTEN/RPA and PTEN/OTUB1 models in PyMol to form a RPA/PTEN/OTUB1 complex model. In this model, the C2 domain of PTEN binds to OTUB1 and the PTEN PTP domain binds to RPA (Figure 4H; PTEN is in cyan, RPA in salmon and OTUB1 in yellow). To verify whether PTEN is necessary for RPA1 and OTUB1 binding, S-tagged pull-down of exogenous Flag-tagged OTUB1 in PTEN+/+ or PTEN−/− HCT116 cells expressing S-tagged RPA1 was carried out. As expected, the recruitment of OTUB1 to RPA1 only occurred in the presence of PTEN (Supplementary information, Figure S4G, lane 2 vs lane 1). Moreover, RPA1 was detected in endogenous OTUB1 immunoprecipitate from PTEN+/+ cells, but was much less detectable in PTEN−/− HCT116 cells (Figure 4I, lane 3 vs lane 4), indicating that PTEN is required for formation of the RPA/PTEN/OTUB1 complex. These findings argue PTEN may recruit OTUB1 to regulate RPA1 deubiquitination. To determine whether PTEN is essential for OTUB1 deubiquitination of RPA1, RPA1 was co-transfected with OTUB1 and/or PTEN into PTEN−/− HCT116 cells. The level of RPA1 ubiquitination was reduced when PTEN or both PTEN and OTUB1 were overexpressed (Supplementary information, Figure S4H, lanes 7, 8 vs lane 5). In contrast, OTUB1 alone had a very limited effect on the level of ubiquitinated RPA1 (Supplementary information, Figure S4H, lane 6). These results suggest that PTEN recruits OTUB1 to deubiquitinate RPA1 in vivo.
To verify the effect of OTUB1 on RPA1 protein stability, TALEN was used to create somatic knockout of OTUB1 in HCT116 cells (Supplementary information, Figure S4I), and endogenous RPA1 protein level in OTUB1+/+ and OTUB1−/− HCT116 cells was evaluated in the presence of CHX (Figure 4J). As expected, homozygous deletion of OTUB1 led to a decrease in expression and stability of endogenous RPA1, similar to PTEN deletion (Figure 4J and 4B). OTUB1+/+ and OTUB1−/− HCT116 cells were then treated with MG132, and the RPA1 expression level was identical in both cell lines (Supplementary information, Figure S4J). In addition, as shown in Figure 4K, RPA1 ubiquitination was elevated in OTUB1−/− HCT116 cells compared with OTUB1+/+ HCT116 cells, suggesting that OTUB1 acts as a DUB for RPA1. DNA fiber length after HU treatment was significantly shorter in OTUB1−/− HCT116 cells than that in control cells (Figure 4L), and RPA1 overexpression compensated for fork instability resulting from OTUB1 deficiency, whereas OTUB1 failed to rescue fork instability following deletion of PTEN or RPA1 (Supplementary information, Figure S4K-S4M). Our results demonstrate that PTEN maintains RPA1 protein stability likely by recruiting OTUB1 to deubiquitinate RPA1.
RPA1 is important for suppression of colorectal carcinoma
As our results suggest a novel tumor suppressor function of PTEN through the regulation of RPA1 in DNA replication, we examined PTEN and RPA1 expression in 20 colorectal cancers and adjacent normal tissues by immunohistochemistry (IHC; Figure 5A). Downregulation of PTEN or RPA1 was observed in 84.2% (16 of 19) or 75% (15 of 20), respectively, of these samples (Figure 5B and 5C), suggesting that both PTEN and RPA1 are downregulated in colorectal cancer cells. In addition, IHC showed wide distribution of PTEN in tumor stroma, but not within tumor cells (Figure 5A). Levels of PTEN and RPA1 showed a significant positive correlation (R = 0.725, P = 0.015) in these colorectal carcinomas (CRC), and 87.5% (14 of 16) of tumors with low PTEN expression also exhibited low RPA1 expression (Figure 5D). These data suggest that PTEN loss may be part of a mechanism that leads to reduction of RPA1 in human CRC.
RPA1 is important for suppression of colorectal carcinoma. (A) Immunohistochemical staining of PTEN and RPA1 in representative colon carcinoma specimens and matched normal colon specimens. Staining (brown) represents positive immunoreactivity. Scale bars, 50 μm. (B, C) PTEN (B) and RPA1 (C) protein expression status in normal colon and colon carcinoma specimens. (D) Correlation of PTEN and RPA1 protein levels in human colon cancers. Statistical significance in B-D was determined with the χ2-test. R is the correlation coefficient. (E) Schematic diagram of strategy for conditional knockout of murine Rpa1. (F) Sketch outlining the AOM/DSS-induced CRC model. Rpa1+/+ and Rpa1+/− mice were treated with or without AOM, which was given once, followed by periodic administration of DSS in water. n = 7 per group. (G-L) Following euthanasia, macroscopic (G) and microscopic (L) analyses of tumor were conducted. Scale bars, 100 μm. Statistical analyses of numbers of tumors (multiplicity, H), numbers of tumors > 3 mm (I), tumor volume (load, J) and average size (K) in two groups.
To evaluate the importance of RPA1 in suppression of colon cancer development, we generated an RPA1 knockout mouse model by homologous recombination. Rpa1flox/flox mice were first created and then crossed with E2A-Cre mice to obtain Rpa1 heterozygous knockout mice (Rpa1+/− mice; Figure 5E). The genome of the Rpa1flox/flox mouse strain was confirmed in targeted ES cells by DNA sequencing, and the genome of E2A-Cre crossed Rpa1+/− mice was confirmed by genotyping and western blotting (Supplementary information, Figure S5A and S5B). No Rpa1−/− mice were obtained, likely due to embryonic lethality. As azoxymethane (AOM) followed by three rounds of dextran sodium sulfate (DSS)29 exposure produces a well-recognized model which mimics the genomic instability in human CRC30,31,32, we generated an AOM/DSS-induced CRC model in Rpa1+/− and Rpa1+/+ mice in vivo as shown in Figure 5F. According to the AOM/DSS protocol (Figure 5F), mice were euthanized and evaluated for cancer. As expected, no tumors were observed in the group without AOM treatment (Figure 5G, Ctl). However, Rpa1+/− mice showed an elevation of tumor multiplicity and tumor load as compared to Rpa1+/+ mice with AOM treatment (Figure 5H-K). Histologic analysis of tumors from both Rpa1+/+ and Rpa1+/− groups showed moderate differentiation with adenomatous patterns, and Rpa1+/− mice developed more poorly differentiated tumors with much higher malignancy (Figure 5L). These results demonstrate that heterozygous disruption of RPA1 in vivo promotes development of CRC.
Discussion
In this study, we demonstrate that PTEN protects stalled DNA replication forks and maintains chromosomal integrity under DNA replication stress. We show that PTEN regulates DNA replication through stabilization of the novel target protein RPA1. PTEN directly binds to RPA1 and may serve as a platform that facilitates OTUB1 deubiquitination and stabilization of RPA1. Expression of PTEN and RPA1 show a significant inverse correlation with CRCs. Our findings represent a novel mechanism through which PTEN regulates DNA replication and suppresses tumorigenesis.
Super resolution images of replication sites obtained using STORM22 reveal the localization of PTEN at replication sites, supporting a function of PTEN in replication regulation. iPOND analysis further demonstrates that PTEN may recruit RPA1 at replication forks. The role of PTEN in replication fork stabilization represents a novel mechanism for tumor suppression. It has been reported that some well known factors such as RAD51, BRCA1/2 and FANCD2 participate in replication fork protection and are associated with the maintenance of genomic integrity20,33. PTEN is able to regulate RAD518, thus it is possible that the PTEN-RPA1 mechanism we describe here is mechanistically linked to the RAD51-BRCA1/2 pathway.
RPA1 is one of the most important eukaryotic single-strand DNA-binding proteins, and serves to protect stalled DNA replication forks. It is also a critical factor in DNA metabolism11,12,34,35. Identification of the functional linkage between PTEN and RPA1 in this study therefore raises the possibility that PTEN is involved in other as yet unidentified aspects of DNA biology. In addition, function of the PTEN-RPA1 pathway in regulation of DNA replication suggests that there is a large network involving these molecules that encompasses additional unidentified mechanisms for obstructing tumorigenesis. One of the important findings in this paper is that loss of RPA1 in mice promotes tumor formation. These findings argue that RPA1 may act as a tumor suppressor in the PTEN signaling pathway.
The physical interaction of the PTEN/RPA1/OTUB1 complex raises the possibility that PTEN may serve as a platform to support interaction of proteins and fine tune their functions. This mechanism should inspire further investigation of potential PTEN function as an essential facilitator for the activities of its other targets. We have recently reported that PTEN can influence the localization of the histone H1 and NPM1 complex on chromatin and regulate chromatin remodeling36. This may in part be analogous to PTEN regulation of RPA1 and OTUB1 function, where the interaction of these molecules brings about deubiquitination and stabilization of RPA1.
This study demonstrates that PTEN protects stalled replication forks and maintains genomic stability through OTUB1-mediated stabilization of RPA1. This novel mechanism provides an unequivocal support to the notion that PTEN is a guardian of the genome.
Materials and Methods
Cell culture, antibodies, and reagents
Primary Pten+/+ and Pten−/− mouse embryonic fibroblasts (MEFs) were previously described8, and NHF3 cells were generated from normal human skin37. HCT116, Hela, and 293T cell lines were obtained from the American Type Culture Collection. PTEN and RPA1 knockdown cell lines were generated by infection with lentivirus followed by puromycin (Sigma) selection. These cells were cultured in DMEM supplemented with 10% FBS in a 37 °C incubator with 5% (v/v) CO2. The insect cell line Sf9 was obtained from Invitrogen and cultured in Grace's insect medium (Gibco).
The following commercial antibodies were used in this study: PTEN (A2B1) and RPA1 (B-6) from Santa Cruz; PTEN (138G6) from Cell Signaling; γ-H2AX (05-636) from Milipore; FLAG (M2-3165) and HA (H3663) from Sigma-Aldrich; GADPH, His, GFP and GST antibodies were from Sungene Biotech (Tianjin, China); β-actin (PM053) was from MBL. Polyclonal antibody to OTUB1 was generated and purified in our laboratory. CHX, CldU, IdU, HU, and MG132 were purchased from Sigma. Cancer tissues and matched adjacent non-tumor tissues were obtained from Peking University 3rd Hospital (Beijing). For the use of these clinical materials for research purposes, prior patient's consent and approval from the Institute Research Ethics Committee were obtained.
Plasmids and shRNA
pSA, pCMV-tag-2b, pGEX-4T-1 and pfastbac1 were purchased from Addgene. Wild-type and mutant PTEN and RPA1 were inserted into these plasmids. pLKO.1-TRC lentiviral shRNA system (Addgene) was used for PTEN and RPA1 knockdown. The target sequences for PTEN shRNA and RPA1 shRNA were 5′-AAAGAGATCGTTAGCAGAA-3′ and 5′-GGCAATCCAGTGCCCTATAAT-3′, respectively, and the scrambled shRNA was against 5′-ACACGTCCGAACATACTAC-3′.
TALEN-mediated somatic RPA1 and OTUB1 knockout
To specifically knockout RPA1 and OTUB1, a TALEN binding pair was chosen for RPA1 or OTUB1 in exon 1 and exon 3, respectively. The genomic recognition sequences of the TALEN left and right arms were 5′-CATGCAGAAGGGGGAT-3′ (L RPA1), 5′-ACGATGACTTGGAGGAT-3′ (R RPA1), and 5′-CTGGTGTCAGAGCGGCTG-3′ (L OTUB1), 5′-CATCTTCAGCATACTCCT-3′ (R OTUB1) spaced by 15 bp and 17 bp, respectively, and anchored by a preceding T base at the −1 position according to optimal criteria for natural TAL proteins38,39. Left and right arm vectors were obtained by one-step ligation using a Fast TALE TM TALEN Assembly Kit (SIDANSAI Biotechnology, China) according to manufacturer's instructions. HCT116 cells were transfected with the TALEN vectors. Puromycin-resistant clones were analyzed by PCR and DNA sequencing to confirm RPA1 or OTUB1 deletions.
Mice and tumor induction
To generate Rpa1flox/− mice, C57BL6J/129S6 hybrid mouse ES cells were electroporated with a targeting vector containing floxed exon 3 and 4. ES cells were selected by G418 and screened for homologous recombination by PCR. Positive ES clones were injected into C57BL/6J blastocysts, and the blastocysts were implanted into pseudopregnant females to generate chimeras. The male chimeras were bred with female Actin-Flpe transgenic mice to delete the FRT-flanked neomycin resistance gene (Neo) from the targeted allele. The neo-deleted Rpa1flox/+ mice were genotyped by PCR. Two heterozygous mice were mated to generate Rpa1flox/flox mice. Rpa1flox/flox mice were backcrossed with C57BL6J for six generations and then mated with E2A-Cre transgenic mice to delete the floxed exon 3 and 4. The targeted Rpa1 allele has an early stop in exon 5, resulting in a truncation of 39 amino acids. Rpa1flox/− mouse was analyzed. Actin-Flpe transgenic mice and E2A-Cre transgenic mice were gifts from Dr Chyuan-Sheng Lin in Columbia University. Both mouse strains were C57BL/6J background. All of the animals were maintained in a special pathogen-free facility at Peking University.
CRC model was induced as described previously30. Briefly, on day 1, mice were injected intraperitoneally with 12.5 mg/kg AOM (National Cancer Institute) and maintained on regular diet and water for 7 days. After 7 days, mice received water with 2.0% dextran sulfate sodium (DSS; MP Biomedicals) for 6 days. Afterword, mice were maintained on regular water for 14 days and subjected to two more DSS treatment cycles. Macroscopic tumors were counted and measured with a caliper. The distal colon was fixed in 10% neutral buffered formalin for over 24 h and transferred to 70% ethanol for subsequent paraffin embedding and histological analysis.
DNA fiber assay
DNA fiber assay was performed as previously described20. Cells were labeled with IdU (50 mM), followed by exposure to HU (4 mM) or were left untreated. DNA fibers were spread as previously described40 prior to standard analysis of IdU or CldU tracts (primary antibodies: IdU, BrdU from BD Biosciences, CldU, BrdU from Novus Biologicals; secondary antibodies: Alexa Fluors 488 and 555, respectively, from Invitrogen). Fibers were imaged (Olympus IX51 microscope) and analyzed using ImageJ software. Statistical analysis was carried out using Prism software.
STORM imaging
Stochastic Optical Reconstruction Microscopy (STORM) imaging was carried out with Nikon N-STORM Super-Resolution Microscope System (LU4A laser box; Nikon Ti-E inverted microscope with Perfect Focus System; Apo TIRF 100× 1.49 Oil; Andor iXon DU897 EMCCD; NIS Elements Software; HP Z820 Work Station). Super-resolution images of Alexa647 (Invitrogen)-labeled DNA replication were acquired at 90 Hz for up to 200 000 frames. The probes were activated by a 405 nm laser (100 mW, Cube, Coherent) at ≤ 100 W/cm2 (at the sample), and the activated Alexa647 molecules were imaged with a 640 nm laser (300 mW, MPB) at ≤ 2 kW/cm2 (at the sample). The super-resolution images of Cy3B (Sigma)-labeled proteins were acquired at 90Hz for up to 200 000 frames under the excitation of a 561 nm laser (150 mW, Coherent) at ≤ 2 kW/cm2 (at the sample). The excitation beams were reflected by a custom-designed polychronic mirror (z405/488/561/640, Chroma). Fluorescence emissions from Atto550 were filtered through a bandpass filter (605/70, Chroma). Ten frames of bright field images were taken for drift correction every second. STORM images were analyzed using Insight3 (provided by Dr Bo Huang) and a custom-written MATLAB script.
CRISPR-mediated BRCA2 knockout cell line
Sequence (GCAGGTTCAGAATTATAGGG) of human BRCA2 gene in exon2 was targeted. Oligos were purchased from Tsingke and ligated into U6-sgRNA plasmid. Human colon cancer cell line HCT116 was seeded onto 6 cm plate (Corning) at a density of 2.5 × 106 cells 24 h prior to transfection. Cells were transfected using PEI at 80%-90% confluence following the manufacturer's recommended protocol. A total of 2 μg Cas9 plasmid and 3 μg of U6-sgRNA plasmid was co-transfected. After G418 selection, cell clones were picked and amplified for mutation sequencing.
Metaphase spreads
For metaphase spreads, cells were seeded and one day later were treated with HU (4 mM), followed by colcemid treatment (0.1 mg/ml) as indicated. Cells were then swollen with 0.075 M KCl (30 min, 37 °C), fixed with methanol/acetic acid (3:1), dropped onto a glass slide, stained with 5% Giemsa, and mounted with Cytoseal 60 (Fisher Scientific) before imaging with an Olympus IX51 microscope.
Chromosomal analysis by C-FISH
Metaphase chromosome spreads were prepared as described above. C-FISH was performed using a Cy3-labeled PNA probe. Both the DNA probes and the slides were heat denatured (80 °C for 5 min) and hybridized at 37 °C for 2 h. Slides were counterstained with DAPI, and the images were captured using an A1+ confocal laser microscope system (Nikon).
Flow cytometry analysis
For cell cycle analysis, cells treated with HU as designed were fixed at −20 °C in 70% (v/v) ethanol overnight, followed by treatment with RNase A (10 mg/ml) at 37 °C for an additional 30 minutes. Cells were resuspended in PBS containing propidium iodide (5 mg/ml) and subjected to flow cytometry. Data acquisition were performed with a FACS Verse apparatus equipped with FACS Diva software (BD Biosciences). Further analysis was carried out using FlowJo software.
S-tag pull-down assay
HCT116 or 293T cells were transfected with pSA, pSA-PTEN, pSA-RPA1, or mutant plasmids and harvested at 24 h after transfection. Cells were lysed in lysis buffer (50 mM Tris pH 7.5, 150 mM NaCl, 2 mM EDTA, 0.5% NP40 and 1 mM NaF). Equal amounts of protein were incubated with s-protein agarose (Novagen) for 4 h. Protein-bead complexes were washed with washing buffer containing 0.5% NP40. Proteins were loaded onto NuPAGE 4%-12% gels (Invitrogen) and visualized with silver staining (Pierce Silver Stain Kit) or subjected to western blot. The potential interacting proteins in specific bands were analyzed with MS analysis.
Immunoprecipitation assay and in vitro binding assay
Immunoprecipitation was performed as previously described8. In brief, cells were extracted and lysed in lysis buffer. Equal amounts of proteins were incubated with antibodies against PTEN, RPA1 or OTUB1 for 4 h, followed by incubation with protein A/G agarose for 1 h. The protein-bead complex mixture was washed in washing buffer containing 0.1% NP40 and subjected to western blot to evaluate protein interaction.
Full-length and truncated forms of PTEN protein were purified using GST beads from E. coli. 2 μg of different GST proteins were first incubated with GST beads for 4 h, followed by a second incubation with in vitro purified His-RPA1 or His-OTUB1 or both proteins from Sf9. Protein-bead complexes were washed in washing buffer containing 0.1% NP-40 for three times and subjected to western blot using an anti-His antibody to detect protein interaction.
The recombinant full-length human PTEN was expressed by pFastbac-PTEN fused with an N-terminal TEV (tobacco etch virus)-cleavable 6-His tag in Sf9 insect cells and was purified by nickel affinity chromatography. Then the affinity tag was removed with TEV protease treatment followed by gel filtration chromatography. PTEN protein was then brought down by immunoprecipitation with the PTEN antibody and incubated with His-OTUB1 and His-RPA1 in the assay.
Chromatin fraction
Cells were lysed in cytoskeleton (CSK) buffer (10 mM PIPES-KOH pH 7.0, 300 mM sucrose, 100 mM NaCl, 3 mM MgCl2, 0.5% Triton X-100) with rotation at 4 °C for 20 min. The pellet after a 5 min centrifugation at 3 000 r.p.m. was washed twice gently with CSK buffer, and resuspended in CSK buffer containing DNase I (RQ1 RNase-Free DNase, Promega, M6101) at a final concentration of 250 U/Pl. The pellet was digested at room temperature for 15 min then at 37 °C for another 15 min, and then centrifuged at 13 000 rpm. for 15 min to obtain the supernatant as the chromatin fraction.
iPOND
iPOND was performed as previously described41,42. PTEN+/+ and PTEN−/− HCT116 cells were labeled with 10 μM EdU for 15 min before 4 mM HU treatment for 5h. Next the cells were crosslinked with 1% formaldehyde for 20 min, permeabilized with 0.25% Triton X-100, washed with 1× PBS, and then incubated for 4 h in click reaction buffer (10 mM Sodium-L-Ascorbate, 10 μM Biotin azide (Glenresearch), 2 mM CuSO4). Cells were resuspended in lysis buffer (1% SDS in 50 mM Tris pH 8.0 containing Roche proteases inhibitor) and sonicated. Samples were centrifuged for 10 min at 16 100× g and supernatants were incubated overnight with streptavidin-coupled beads (Calbiochem). An aliquot (1%) of the extract was kept as loading control. The beads were washed in lysis buffer and 1 M NaCl, and then incubated in 2× SDS sample buffer for 30 min at 95 °C.
In vivo ubiquitination and deubiquitination assay
PTEN+/+, PTEN−/− and OTUB1+/+, OTUB1−/− HCT116 cells were transfected with Flag-RPA1 or control and His-HA-ubiquitin. 24 h after transfection, cells were treated with 10 μM MG132 for 10 h. Cells were collected and protein extract was prepared, then in vivo ubiquitination assay was performed as described previously43. The ubiquitination ladder was detected with an anti-Flag antibody.
For the in vivo deubiquitination assay, PTEN−/− HCT116 cells were transfected with Flag-RPA1 or control, His-HA–ubiquitin, S-HA-PTEN and/or S-HA-OTUB1 for 24 h, then treated with 10 μM MG132 for 6 h. His-HA-Ubiquitin-Flag-RPA1 was pulled-down with Ni-beads and subjected to immunoblotting with an antibody against Flag.
Confocal microscopy
For evaluation of endogenous co-localization, PTEN+/+ and PTEN−/− HCT116 cells were seeded on glass cover slips in DMEM medium for 24 h. Cells were fixed with acetone for 10 min, followed by blocking with 1% BSA and incubation with specific primary antibodies at 4 °C overnight. After incubation with secondary antibodies (Alexa Fluors 488 and 555, Invitrogen) at room temperature for 1 h and staining with 0.5 μg/ml DAPI for 10 min, cover glasses were mounted and evaluated with fluorescence microscopy. A Nikon TCS A1 microscope was used for confocal microscopy.
Immunohistochemistry
Samples were deparaffinized and rehydrated. Endogenous peroxidase activity was blocked with 3% (v/v) hydrogen peroxide in methanol for 10 min. Antigen retrieval was carried out using 1 mM EDTA buffer (PH 9.0) in a microwave oven. After 1 h pre-incubation in 5% BSA to prevent nonspecific staining, samples were incubated with antibodies against PTEN (1:200, Cell Signaling) and RPA1 (1:100, Santa Cruz) at 4 °C overnight. Sections were developed with the Envision Detection System (Dako) and counterstained with haematoxylin. Images were acquired using an Olympus IX51 microscope. From both the percentage of immunopositive cells and immunostaining intensity, a total protein expression score was calculated. High and low protein expression was defined using the mean score of all samples as a cutoff value. The χ2-test was used for statistical analysis of correlation of PTEN or RPA1 with tissue type, and correlation of PTEN and RPA1.
In silico docking
For modeling of the PTEN/RPA and PTEN/OTUB1 complexes, we separately submitted the crystal structure of PTEN (PDB:1D5R) as a receptor and RPARPA1(439-616)/RPA2(43-171)/RPA323 or OTUB1 (PDB: 2ZFY)28 as ligand to the web-based ClusPro2.0 docking server (http://cluspro.bu.edu/). This docking program filters docked conformations with good surface and charge complementarity and ranks them based on their clustering properties25. Then we merged PTEN/RPA and PTEN/OTUB1 model in PyMol to get a RPA/PTEN/OTUB1 complex model. All docking trials were carried out using balanced mode with default parameters. Output images were generated in PyMol.
Statistical analysis
The densitometric quantification of western blot levels was performed using Quantity One software. Prism GraphPad software v5.01 was used for statistical analysis. The statistical significance of differences between various groups was calculated with the two-tailed unpaired t-test, and error bars represent SEM. P-values of 0.05 or less were considered statistically significant.
References
Aguilera A, Gomez-Gonzalez B . Genome instability: a mechanistic view of its causes and consequences. Nat Rev Genet 2008; 9:204–217.
Branzei D, Foiani M . Maintaining genome stability at the replication fork. Nat Rev Mol Cell Biol 2010; 11:208–219.
Cimprich MKZaKA. Causes and consequences of replication stress. Nat Cell Biol 2014; 16:2–9.
Negrini S, Gorgoulis VG, Halazonetis TD . Genomic instability--an evolving hallmark of cancer. Nat Rev Mol Cell Biol 2010; 11:220–228.
Biankin AV, Waddell N, Kassahn KS, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature 2012; 491:399–405.
Di Cristofano A, Pandolfi PP . The multiple roles of PTEN in tumor suppression. Cell 2000; 100:387–390.
Salmena L, Carracedo A, Pandolfi PP . Tenets of PTEN tumor suppression. Cell 2008; 133:403–414.
Shen WH, Balajee AS, Wang J, et al. Essential role for nuclear PTEN in maintaining chromosomal integrity. Cell 2007; 128:157–170.
Song MS, Salmena L, Pandolfi PP . The functions and regulation of the PTEN tumour suppressor. Nat Rev Mol Cell Biol 2012; 13:283–296.
Sun Z, Huang C, He J, et al. PTEN C-terminal deletion causes genomic instability and tumor development. Cell Rep 2014; 6:844–854.
Fanning E, Klimovich V, Nager AR . A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res 2006; 34:4126–4137.
Zou Y, Liu Y, Wu X, Shell SM . Functions of human replication protein A (RPA): from DNA replication to DNA damage and stress responses. J Cell Physiol 2006; 208:267–273.
Yuan J, Ghosal G, Chen J . The annealing helicase HARP protects stalled replication forks. Genes Dev 2009; 23:2394–2399.
Toledo LI, Altmeyer M, Rask MB, et al. ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell 2013; 155:1088–1103.
Mevissen TE, Hospenthal MK, Geurink PP, et al. OTU deubiquitinases reveal mechanisms of linkage specificity and enable ubiquitin chain restriction analysis. Cell 2013; 154:169–184.
Wiener R, Zhang X, Wang T, Wolberger C . The mechanism of OTUB1-mediated inhibition of ubiquitination. Nature 2012; 483:618–622.
Wiener R, DiBello AT, Lombardi PM, et al. E2 ubiquitin-conjugating enzymes regulate the deubiquitinating activity of OTUB1. Nat Struct Mol Biol 2013; 20:1033–1039.
Juang YC, Landry MC, Sanches M, et al. OTUB1 co-opts Lys48-linked ubiquitin recognition to suppress E2 enzyme function. Mol Cell 2012; 45:384–397.
Balakirev MY, Tcherniuk SO, Jaquinod M, Chroboczek J . Otubains: a new family of cysteine proteases in the ubiquitin pathway. EMBO Rep 2003; 4:517–522.
Schlacher K, Christ N, Siaud N, Egashira A, Wu H, Jasin M . Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell 2011; 145:529–542.
Vlahos CJ, Matter WF, Hui KY, Brown RF . A specific inhibitor of phosphatidylinositol 3-kinase, 2-(4-morpholinyl)-8-phenyl-4H-1-benzopyran-4-one (LY294002). J Biol Chem 1994; 269:5241–5248.
Rust MJ, Bates M, Zhuang X . Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat Methods 2006; 3:793–795.
Bochkareva E, Korolev S, Lees-Miller SP, Bochkarev A . Structure of the RPA trimerization core and its role in the multistep DNA-binding mechanism of RPA. EMBO J 2002; 21:1855–1863.
Lee JO, Yang H, Georgescu MM, et al. Crystal structure of the PTEN tumor suppressor: implications for its phosphoinositide phosphatase activity and membrane association. Cell 1999; 99:323–334.
Kozakov D, Hall DR, Beglov D, et al. Achieving reliability and high accuracy in automated protein docking: ClusPro, PIPER, SDU, and stability analysis in CAPRI rounds 13-19. Proteins 2010; 78:3124–3130.
Dou H, Huang C, Singh M, Carpenter PB, Yeh ET . Regulation of DNA repair through deSUMOylation and SUMOylation of replication protein A complex. Mol Cell 2010; 39:333–345.
Marechal A, Li JM, Ji XY, et al. PRP19 Transforms into a sensor of RPA-ssDNA after DNA damage and drives ATR activation via a ubiquitin-mediated circuitry. Mol Cell 2013; 53:235–246.
Edelmann MJ, Iphofer A, Akutsu M, et al. Structural basis and specificity of human otubain 1-mediated deubiquitination. Biochem J 2009; 418:379–390.
Stacey SN, Gudbjartsson DF, Sulem P, et al. Common variants on 1p36 and 1q42 are associated with cutaneous basal cell carcinoma but not with melanoma or pigmentation traits. Nat Genet 2008; 40:1313–1318.
Greten FR, Eckmann L, Greten TF, et al. IKKbeta links inflammation and tumorigenesis in a mouse model of colitis-associated cancer. Cell 2004; 118:285–296.
Grivennikov S, Karin E, Terzic J, et al. IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell 2009; 15:103–113.
Valeri N, Braconi C, Gasparini P, et al. MicroRNA-135b promotes cancer progression by acting as a downstream effector of oncogenic pathways in colon cancer. Cancer Cell 2014; 25:469–483.
Schlacher K, Wu H, Jasin M . A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell 2012; 22:106–116.
Flynn RL, Centore RC, O'Sullivan RJ, et al. TERRA and hnRNPA1 orchestrate an RPA-to-POT1 switch on telomeric single-stranded DNA. Nature 2011; 471:532–536.
Fujimoto M, Takaki E, Takii R, et al. RPA assists HSF1 access to nucleosomal DNA by recruiting histone chaperone FACT. Mol Cell 2012; 48:182–194.
Chen ZH, Zhu M, Yang J, et al. PTEN interacts with histone H1 and controls chromatin condensation. Cell Rep 2014; 8:2003–2014.
Yin Y, Terauchi Y, Solomon GG, et al. Involvement of p85 in p53-dependent apoptotic response to oxidative stress. Nature 1998; 391:707–710.
Boch J, Scholze H, Schornack S, et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science 2009; 326:1509–1512.
Moscou MJ, Bogdanove, AJ . A simple cipher governs DNA recognition by TAL effectors. Science 2009; 326.
Jackson DA, Pombo A . Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J Cell Biol 1998; 140:1285–1295.
Sirbu BM, Couch FB, Cortez D . Monitoring the spatiotemporal dynamics of proteins at replication forks and in assembled chromatin using isolation of proteins on nascent DNA. Nat Protoc 2012; 7:594–605.
Lossaint G, Larroque M, Ribeyre C, et al. FANCD2 binds MCM proteins and controls replisome function upon activation of s phase checkpoint signaling. Mol Cell 2013; 51:678–690.
Li M, Brooks CL, Wu-Baer F, Chen D, Baer R, Gu W . Mono- versus polyubiquitination: differential control of p53 fate by Mdm2. Science 2003; 302:1972–1975.
Acknowledgements
We thank L Liang for solving the in silico docking analysis and protein purification, C Song for generating the BRCA2+/− HCT116 cell line, X Du, C Zhang and Y Liu for data analysis and manuscript preparation, Y Li for OTUB1 antibody generation. We thank Dr Chyuan-Sheng Lin for Actin-Flpe and E2A-Cre transgenic mice. We thank J Liang, X Zhao, Q Yin, X Chen, S He, J Zhou, and W Zhao for technical assistance and critical discussions. This study was supported by grants to YY including the National Natural Scientific Foundation of China (NSFC 31420103905, 81430056, 81372491 and 81321003), the China National Major Scientific Program (2010CB912202), the Shu Fan Education and Research Foundation, and Lam Chung Nin Foundation for Systems Biomedicine.
Author information
Authors and Affiliations
Corresponding author
Additional information
( Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary information
Supplementary information, Figure S1
PTEN regulates DNA replication mainly through stalled replication forks protection (PDF 385 kb)
Supplementary information, Figure S2
PTEN binds to RPA1 mainly through its PTP domain (PDF 338 kb)
Supplementary information, Figure S3
RPA1 is required for stalled replication forks protection (PDF 264 kb)
Supplementary information, Figure S4
RPA1 is stabilized by PTEN/OTUB1 and acts as a downstream target (PDF 414 kb)
Supplementary information, Figure S5
Validation of Rpa1+/− mice (PDF 310 kb)
Rights and permissions
About this article
Cite this article
Wang, G., Li, Y., Wang, P. et al. PTEN regulates RPA1 and protects DNA replication forks. Cell Res 25, 1189–1204 (2015). https://doi.org/10.1038/cr.2015.115
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cr.2015.115
Keywords
This article is cited by
-
LAP2α preserves genome integrity through assisting RPA deposition on damaged chromatin
Genome Biology (2022)
-
The equilibrium of tumor suppression: DUBs as active regulators of PTEN
Experimental & Molecular Medicine (2022)
-
Decoding the pathogenesis of Diamond–Blackfan anemia using single-cell RNA-seq
Cell Discovery (2022)
-
Depletion of NK6 Homeobox 3 (NKX6.3) causes gastric carcinogenesis through copy number alterations by inducing impairment of DNA replication and repair regulation
Oncogenesis (2021)
-
The phosphatase PAC1 acts as a T cell suppressor and attenuates host antitumor immunity
Nature Immunology (2020)